









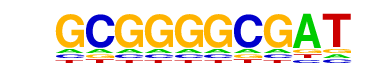


| p-value: | 1e-446 |
| log p-value: | -1.027e+03 |
| Information Content per bp: | 1.886 |
| Number of Target Sequences with motif | 503.0 |
| Percentage of Target Sequences with motif | 9.14% |
| Number of Background Sequences with motif | 17.0 |
| Percentage of Background Sequences with motif | 0.51% |
| Average Position of motif in Targets | 102.1 +/- 52.4bp |
| Average Position of motif in Background | 116.2 +/- 60.9bp |
| Strand Bias (log2 ratio + to - strand density) | 0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.22 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.894 |  | 1e-233 | -537.028158 | 12.77% | 2.89% | motif file (matrix) |
| 2 | 0.886 |  | 1e-173 | -398.922465 | 11.74% | 3.20% | motif file (matrix) |
| 3 | 0.716 |  | 1e-159 | -367.983232 | 9.01% | 2.06% | motif file (matrix) |
| 4 | 0.765 |  | 1e-144 | -333.432486 | 2.53% | 0.11% | motif file (matrix) |
| 5 | 0.883 |  | 1e-138 | -319.185207 | 3.49% | 0.28% | motif file (matrix) |
| 6 | 0.619 |  | 1e-138 | -318.109834 | 2.18% | 0.08% | motif file (matrix) |
| 7 | 0.648 |  | 1e-124 | -286.796361 | 1.69% | 0.04% | motif file (matrix) |
| 8 | 0.628 |  | 1e-121 | -278.726316 | 1.65% | 0.05% | motif file (matrix) |
| 9 | 0.721 |  | 1e-109 | -252.854529 | 2.27% | 0.14% | motif file (matrix) |
| 10 | 0.757 |  | 1e-106 | -245.023071 | 6.67% | 1.67% | motif file (matrix) |
| 11 | 0.662 |  | 1e-102 | -235.162679 | 1.45% | 0.03% | motif file (matrix) |