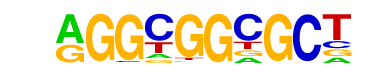







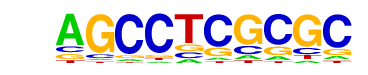





| p-value: | 1e-422 |
| log p-value: | -9.718e+02 |
| Information Content per bp: | 1.835 |
| Number of Target Sequences with motif | 363.0 |
| Percentage of Target Sequences with motif | 9.52% |
| Number of Background Sequences with motif | 6.9 |
| Percentage of Background Sequences with motif | 0.30% |
| Average Position of motif in Targets | 97.5 +/- 51.8bp |
| Average Position of motif in Background | 54.4 +/- 31.1bp |
| Strand Bias (log2 ratio + to - strand density) | -0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.20 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.788 |  | 1e-188 | -434.883400 | 3.36% | 0.04% | motif file (matrix) |
| 2 | 0.908 |  | 1e-179 | -412.762796 | 16.49% | 4.34% | motif file (matrix) |
| 3 | 0.721 |  | 1e-140 | -323.931850 | 2.67% | 0.08% | motif file (matrix) |
| 4 | 0.627 |  | 1e-139 | -322.155914 | 3.17% | 0.13% | motif file (matrix) |
| 5 | 0.695 |  | 1e-134 | -309.818583 | 4.04% | 0.26% | motif file (matrix) |
| 6 | 0.668 |  | 1e-129 | -298.703273 | 3.67% | 0.19% | motif file (matrix) |
| 7 | 0.707 |  | 1e-124 | -286.102801 | 5.22% | 0.55% | motif file (matrix) |
| 8 | 0.677 |  | 1e-101 | -232.721600 | 3.09% | 0.18% | motif file (matrix) |
| 9 | 0.608 |  | 1e-88 | -204.547542 | 1.89% | 0.08% | motif file (matrix) |
| 10 | 0.602 |  | 1e-72 | -167.678116 | 2.25% | 0.16% | motif file (matrix) |
| 11 | 0.770 |  | 1e-67 | -156.509561 | 1.55% | 0.08% | motif file (matrix) |
| 12 | 0.671 |  | 1e-60 | -138.771255 | 1.42% | 0.04% | motif file (matrix) |