| p-value: | 1e-77 |
| log p-value: | -1.784e+02 |
| Information Content per bp: | 1.825 |
| Number of Target Sequences with motif | 65.0 |
| Percentage of Target Sequences with motif | 1.70% |
| Number of Background Sequences with motif | 1.7 |
| Percentage of Background Sequences with motif | 0.07% |
| Average Position of motif in Targets | 99.0 +/- 56.5bp |
| Average Position of motif in Background | 125.9 +/- 14.5bp |
| Strand Bias (log2 ratio + to - strand density) | -0.5 |
| Multiplicity (# of sites on avg that occur together) | 1.03 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
Rbpj1(?)/Panc1-Rbpj1-ChIP-Seq(GSE47459)/Homer
| Match Rank: | 1 |
| Score: | 0.67
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --TTCCGAGGCG
HTTTCCCASG-- |
|


|
|
MA0154.2_EBF1/Jaspar
| Match Rank: | 2 |
| Score: | 0.59
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | TTCCGAGGCG--
-TCCCTGGGGAN |
|


|
|
PB0115.1_Ehf_2/Jaspar
| Match Rank: | 3 |
| Score: | 0.58
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------TTCCGAGGCG
TAGTATTTCCGATCTT |
|


|
|
MA0028.1_ELK1/Jaspar
| Match Rank: | 4 |
| Score: | 0.58
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TTCCGAGGCG
CTTCCGGNNN- |
|


|
|
Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 5 |
| Score: | 0.58
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---TTCCGAGGCG
NRYTTCCGGY--- |
|


|
|
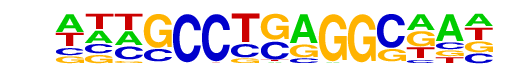
PB0088.1_Tcfap2e_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.57
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---TTCCGAGGCG--
ATTGCCTGAGGCAAT |
|

|
|
Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 7 |
| Score: | 0.57
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---TTCCGAGGCG
HACTTCCGGY--- |
|


|
|
EBF1(EBF)/Near-E2A-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 8 |
| Score: | 0.57
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -TTCCGAGGCG-
GTCCCCAGGGGA |
|


|
|
ZNF711(Zf)/SH-SY5Y-ZNF711-ChIP-Seq(GSE20673)/Homer
| Match Rank: | 9 |
| Score: | 0.56
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | TTCCGAGGCG-
---CTAGGCCT |
|


|
|
PH0048.1_Hoxa13/Jaspar
| Match Rank: | 10 |
| Score: | 0.55
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----TTCCGAGGCG-
ANATTTTACGAGNNNN |
|


|
|





















