| p-value: | 1e-14 |
| log p-value: | -3.453e+01 |
| Information Content per bp: | 1.693 |
| Number of Target Sequences with motif | 107.0 |
| Percentage of Target Sequences with motif | 1.94% |
| Number of Background Sequences with motif | 355.1 |
| Percentage of Background Sequences with motif | 0.81% |
| Average Position of motif in Targets | 98.0 +/- 52.2bp |
| Average Position of motif in Background | 94.5 +/- 60.7bp |
| Strand Bias (log2 ratio + to - strand density) | 0.5 |
| Multiplicity (# of sites on avg that occur together) | 1.01 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
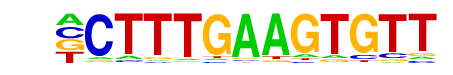
TCFL2(HMG)/K562-TCF7L2-ChIP-Seq(GSE29196)/Homer
| Match Rank: | 1 |
| Score: | 0.81
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CTTTGAAGTGTT
CCTTTGAWGT--- |
|

|
|
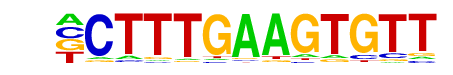
Tcf3(HMG)/mES-Tcf3-ChIP-Seq(GSE11724)/Homer
| Match Rank: | 2 |
| Score: | 0.81
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CTTTGAAGTGTT
CCTTTGATGT--- |
|

|
|
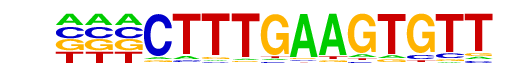
MA0523.1_TCF7L2/Jaspar
| Match Rank: | 3 |
| Score: | 0.78
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---CTTTGAAGTGTT
TNCCTTTGATCTTN- |
|

|
|
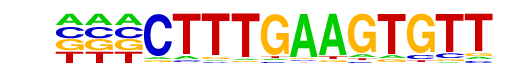
Tcf4(HMG)/Hct116-Tcf4-ChIP-Seq(SRA012054)/Homer
| Match Rank: | 4 |
| Score: | 0.76
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---CTTTGAAGTGTT
TNCCTTTGATGT--- |
|

|
|
PB0082.1_Tcf3_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.71
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----CTTTGAAGTGTT
NNTTCCTTTGATCTANA |
|


|
|
PB0083.1_Tcf7_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.69
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----CTTTGAAGTGTT
NNTTCCTTTGATCTNNA |
|


|
|
PB0084.1_Tcf7l2_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.69
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----CTTTGAAGTGTT
ATTTCCTTTGATCTATA |
|


|
|
PH0111.1_Nkx2-2/Jaspar
| Match Rank: | 8 |
| Score: | 0.68
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CTTTGAAGTGTT---
NANTTTCAAGTGGTTAN |
|


|
|
PB0040.1_Lef1_1/Jaspar
| Match Rank: | 9 |
| Score: | 0.67
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----CTTTGAAGTGTT
AATCCCTTTGATCTATC |
|


|
|
Nkx2.1(Homeobox)/LungAC-Nkx2.1-ChIP-Seq(GSE43252)/Homer
| Match Rank: | 10 |
| Score: | 0.63
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CTTTGAAGTGTT
--CTYRAGTGSY |
|


|
|





















