


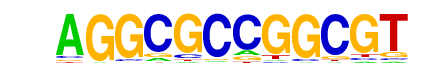


















| p-value: | 1e-373 |
| log p-value: | -8.598e+02 |
| Information Content per bp: | 1.734 |
| Number of Target Sequences with motif | 219.0 |
| Percentage of Target Sequences with motif | 5.74% |
| Number of Background Sequences with motif | 1.8 |
| Percentage of Background Sequences with motif | 0.08% |
| Average Position of motif in Targets | 105.9 +/- 51.2bp |
| Average Position of motif in Background | 94.9 +/- 46.5bp |
| Strand Bias (log2 ratio + to - strand density) | -0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.13 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
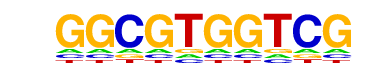
| 1 | 0.763 |  | 1e-310 | -714.010873 | 4.95% | 0.08% | motif file (matrix) |
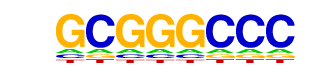
| 2 | 0.803 |  | 1e-255 | -588.678874 | 4.98% | 0.11% | motif file (matrix) |
| 3 | 0.657 |  | 1e-223 | -514.136988 | 11.48% | 1.62% | motif file (matrix) |
| 4 | 0.684 |  | 1e-196 | -451.666426 | 3.46% | 0.04% | motif file (matrix) |
| 5 | 0.624 |  | 1e-194 | -447.265107 | 3.43% | 0.05% | motif file (matrix) |
| 6 | 0.679 |  | 1e-180 | -414.482867 | 3.83% | 0.11% | motif file (matrix) |
| 7 | 0.728 |  | 1e-168 | -388.342076 | 7.52% | 0.87% | motif file (matrix) |
| 8 | 0.683 |  | 1e-166 | -384.186317 | 3.62% | 0.11% | motif file (matrix) |
| 9 | 0.767 |  | 1e-159 | -368.226532 | 6.47% | 0.65% | motif file (matrix) |
| 10 | 0.747 |  | 1e-157 | -362.823640 | 7.81% | 1.02% | motif file (matrix) |
| 11 | 0.666 |  | 1e-142 | -327.731957 | 4.69% | 0.33% | motif file (matrix) |
| 12 | 0.681 |  | 1e-141 | -326.766863 | 4.19% | 0.24% | motif file (matrix) |
| 13 | 0.662 |  | 1e-136 | -315.055728 | 2.62% | 0.08% | motif file (matrix) |
| 14 | 0.633 |  | 1e-124 | -285.932390 | 4.69% | 0.40% | motif file (matrix) |
| 15 | 0.604 |  | 1e-116 | -268.249310 | 3.67% | 0.24% | motif file (matrix) |
| 16 | 0.727 |  | 1e-115 | -265.174361 | 6.16% | 0.88% | motif file (matrix) |
| 17 | 0.682 |  | 1e-108 | -250.513024 | 2.20% | 0.09% | motif file (matrix) |
| 18 | 0.616 |  | 1e-98 | -226.282021 | 3.28% | 0.23% | motif file (matrix) |
| 19 | 0.681 |  | 1e-96 | -223.190337 | 3.46% | 0.31% | motif file (matrix) |
| 20 | 0.634 |  | 1e-95 | -219.679488 | 5.29% | 0.82% | motif file (matrix) |
| 21 | 0.792 |  | 1e-37 | -86.168161 | 1.26% | 0.11% | motif file (matrix) |
| 22 | 0.691 |  | 1e-34 | -80.156168 | 3.77% | 1.14% | motif file (matrix) |