| p-value: | 1e-14 |
| log p-value: | -3.273e+01 |
| Information Content per bp: | 1.707 |
| Number of Target Sequences with motif | 80.0 |
| Percentage of Target Sequences with motif | 2.10% |
| Number of Background Sequences with motif | 352.0 |
| Percentage of Background Sequences with motif | 0.77% |
| Average Position of motif in Targets | 112.6 +/- 52.5bp |
| Average Position of motif in Background | 87.2 +/- 58.3bp |
| Strand Bias (log2 ratio + to - strand density) | 0.5 |
| Multiplicity (# of sites on avg that occur together) | 1.05 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
GFY(?)/Promoter/Homer
| Match Rank: | 1 |
| Score: | 0.95
| | Offset: | 0
| | Orientation: | forward strand |
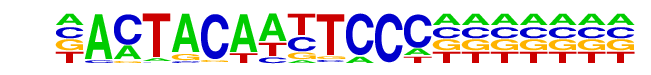
| Alignment: | ACTACAAYTCCC
ACTACAATTCCC |
|


|
|
GFY-Staf/Promoters/Homer
| Match Rank: | 2 |
| Score: | 0.81
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -ACTACAAYTCCC-------
AACTACAATTCCCAGAATGC |
|

|
|
PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 3 |
| Score: | 0.56
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | ACTACAAYTCCC--
----CACTTCCTCT |
|


|
|
MA0056.1_MZF1_1-4/Jaspar
| Match Rank: | 4 |
| Score: | 0.55
| | Offset: | 8
| | Orientation: | reverse strand |
| Alignment: | ACTACAAYTCCC--
--------TCCCCA |
|


|
|
MA0019.1_Ddit3::Cebpa/Jaspar
| Match Rank: | 5 |
| Score: | 0.54
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -ACTACAAYTCCC
AGATGCAATCCC- |
|


|
|
MA0156.1_FEV/Jaspar
| Match Rank: | 6 |
| Score: | 0.54
| | Offset: | 5
| | Orientation: | reverse strand |
| Alignment: | ACTACAAYTCCC-
-----ATTTCCTG |
|


|
|
ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer
| Match Rank: | 7 |
| Score: | 0.53
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | ACTACAAYTCCC--
----CACTTCCTGT |
|


|
|
MA0136.1_ELF5/Jaspar
| Match Rank: | 8 |
| Score: | 0.52
| | Offset: | 4
| | Orientation: | forward strand |
| Alignment: | ACTACAAYTCCC-
----TACTTCCTT |
|


|
|
PB0091.1_Zbtb3_1/Jaspar
| Match Rank: | 9 |
| Score: | 0.52
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------ACTACAAYTCCC
AATCGCACTGCATTCCG- |
|


|
|
MA0511.1_RUNX2/Jaspar
| Match Rank: | 10 |
| Score: | 0.52
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---ACTACAAYTCCC
CAAACCACAAACCCC |
|


|
|





















