

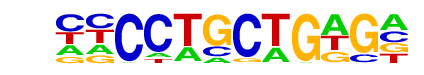






| p-value: | 1e-290 |
| log p-value: | -6.699e+02 |
| Information Content per bp: | 1.669 |
| Number of Target Sequences with motif | 925.0 |
| Percentage of Target Sequences with motif | 8.76% |
| Number of Background Sequences with motif | 793.9 |
| Percentage of Background Sequences with motif | 2.03% |
| Average Position of motif in Targets | 98.3 +/- 48.4bp |
| Average Position of motif in Background | 101.5 +/- 59.7bp |
| Strand Bias (log2 ratio + to - strand density) | -0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.03 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.906 |  | 1e-199 | -459.748185 | 22.60% | 12.06% | motif file (matrix) |
| 2 | 0.628 |  | 1e-176 | -405.613311 | 26.83% | 15.96% | motif file (matrix) |
| 3 | 0.645 |  | 1e-168 | -386.855269 | 22.94% | 13.06% | motif file (matrix) |
| 4 | 0.660 |  | 1e-126 | -291.423392 | 37.82% | 27.09% | motif file (matrix) |
| 5 | 0.720 |  | 1e-88 | -204.585537 | 15.25% | 9.15% | motif file (matrix) |
| 6 | 0.730 |  | 1e-46 | -107.160944 | 8.39% | 5.05% | motif file (matrix) |
| 7 | 0.680 |  | 1e-27 | -62.930132 | 3.37% | 1.79% | motif file (matrix) |