| p-value: | 1e-7 |
| log p-value: | -1.776e+01 |
| Information Content per bp: | 1.724 |
| Number of Target Sequences with motif | 13.0 |
| Percentage of Target Sequences with motif | 0.12% |
| Number of Background Sequences with motif | 6.2 |
| Percentage of Background Sequences with motif | 0.02% |
| Average Position of motif in Targets | 118.8 +/- 63.5bp |
| Average Position of motif in Background | 90.7 +/- 33.2bp |
| Strand Bias (log2 ratio + to - strand density) | 0.7 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PB0030.1_Hnf4a_1/Jaspar
| Match Rank: | 1 |
| Score: | 0.64
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---ACGGGGTCCATC--
CTCCAGGGGTCAATTGA |
|


|
|
PB0025.1_Glis2_1/Jaspar
| Match Rank: | 2 |
| Score: | 0.56
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----ACGGGGTCCATC
NTNTGGGGGGTCNNNA |
|


|
|
PB0156.1_Plagl1_2/Jaspar
| Match Rank: | 3 |
| Score: | 0.56
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---ACGGGGTCCATC--
GCTGGGGGGTACCCCTT |
|


|
|
PB0057.1_Rxra_1/Jaspar
| Match Rank: | 4 |
| Score: | 0.55
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---ACGGGGTCCATC--
NTNNNGGGGTCANGNNN |
|


|
|
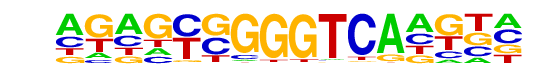
PB0157.1_Rara_2/Jaspar
| Match Rank: | 5 |
| Score: | 0.54
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---ACGGGGTCCATC-
AGAGCGGGGTCAAGTA |
|

|
|
PB0153.1_Nr2f2_2/Jaspar
| Match Rank: | 6 |
| Score: | 0.53
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---ACGGGGTCCATC-
CGCGCCGGGTCACGTA |
|


|
|
PB0118.1_Esrra_2/Jaspar
| Match Rank: | 7 |
| Score: | 0.51
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---ACGGGGTCCATC--
GGCGAGGGGTCAAGGGC |
|


|
|
PB0052.1_Plagl1_1/Jaspar
| Match Rank: | 8 |
| Score: | 0.50
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --ACGGGGTCCATC--
NNNGGGGCGCCCCCNN |
|


|
|
MA0059.1_MYC::MAX/Jaspar
| Match Rank: | 9 |
| Score: | 0.49
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----ACGGGGTCCATC
GACCACGTGGT----- |
|


|
|
MA0057.1_MZF1_5-13/Jaspar
| Match Rank: | 10 |
| Score: | 0.49
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --ACGGGGTCCATC
GGAGGGGGAA---- |
|


|
|





















