





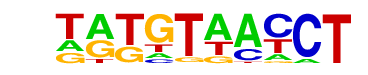




| p-value: | 1e-58 |
| log p-value: | -1.350e+02 |
| Information Content per bp: | 1.666 |
| Number of Target Sequences with motif | 261.0 |
| Percentage of Target Sequences with motif | 15.56% |
| Number of Background Sequences with motif | 2364.1 |
| Percentage of Background Sequences with motif | 4.94% |
| Average Position of motif in Targets | 109.3 +/- 53.9bp |
| Average Position of motif in Background | 100.0 +/- 60.1bp |
| Strand Bias (log2 ratio + to - strand density) | -0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.16 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.907 |  | 1e-36 | -83.175060 | 10.20% | 3.34% | motif file (matrix) |
| 2 | 0.711 |  | 1e-23 | -54.812378 | 1.13% | 0.03% | motif file (matrix) |
| 3 | 0.832 |  | 1e-23 | -53.345075 | 3.22% | 0.56% | motif file (matrix) |
| 4 | 0.621 |  | 1e-21 | -48.994615 | 10.08% | 4.47% | motif file (matrix) |
| 5 | 0.687 |  | 1e-18 | -42.344597 | 22.06% | 14.02% | motif file (matrix) |
| 6 | 0.698 |  | 1e-14 | -33.679514 | 0.95% | 0.05% | motif file (matrix) |
| 7 | 0.638 |  | 1e-12 | -27.852250 | 14.07% | 8.79% | motif file (matrix) |
| 8 | 0.692 |  | 1e-11 | -26.452068 | 61.42% | 53.07% | motif file (matrix) |
| 9 | 0.733 |  | 1e-9 | -20.883887 | 0.42% | 0.01% | motif file (matrix) |