





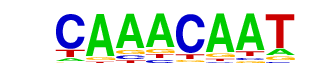







| p-value: | 1e-285 |
| log p-value: | -6.566e+02 |
| Information Content per bp: | 1.706 |
| Number of Target Sequences with motif | 1713.0 |
| Percentage of Target Sequences with motif | 23.82% |
| Number of Background Sequences with motif | 3950.9 |
| Percentage of Background Sequences with motif | 9.32% |
| Average Position of motif in Targets | 99.8 +/- 51.6bp |
| Average Position of motif in Background | 100.2 +/- 59.0bp |
| Strand Bias (log2 ratio + to - strand density) | -0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.17 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.919 |  | 1e-278 | -641.703858 | 29.30% | 13.19% | motif file (matrix) |
| 2 | 0.895 |  | 1e-263 | -606.204460 | 33.29% | 16.51% | motif file (matrix) |
| 3 | 0.874 |  | 1e-174 | -401.526273 | 19.02% | 8.43% | motif file (matrix) |
| 4 | 0.712 |  | 1e-116 | -268.536318 | 16.95% | 8.47% | motif file (matrix) |
| 5 | 0.714 |  | 1e-105 | -243.707915 | 23.41% | 13.78% | motif file (matrix) |
| 6 | 0.795 |  | 1e-101 | -233.102471 | 17.37% | 9.26% | motif file (matrix) |
| 7 | 0.711 |  | 1e-93 | -215.979551 | 31.65% | 21.21% | motif file (matrix) |
| 8 | 0.750 |  | 1e-61 | -142.312447 | 11.14% | 5.96% | motif file (matrix) |
| 9 | 0.609 |  | 1e-57 | -132.425971 | 12.14% | 6.87% | motif file (matrix) |
| 10 | 0.644 |  | 1e-41 | -95.535724 | 10.11% | 5.97% | motif file (matrix) |
| 11 | 0.648 |  | 1e-28 | -64.650634 | 4.37% | 2.20% | motif file (matrix) |
| 12 | 0.633 |  | 1e-11 | -25.600124 | 4.51% | 3.04% | motif file (matrix) |