| p-value: | 1e-7 |
| log p-value: | -1.731e+01 |
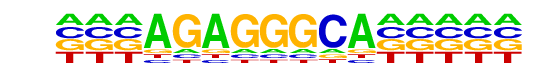
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 178.0 |
| Percentage of Target Sequences with motif | 2.47% |
| Number of Background Sequences with motif | 680.8 |
| Percentage of Background Sequences with motif | 1.61% |
| Average Position of motif in Targets | 100.1 +/- 52.3bp |
| Average Position of motif in Background | 97.3 +/- 60.0bp |
| Strand Bias (log2 ratio + to - strand density) | 0.4 |
| Multiplicity (# of sites on avg that occur together) | 1.02 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
MA0512.1_Rxra/Jaspar
| Match Rank: | 1 |
| Score: | 0.75
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -AGAGGGCA--
CAAAGGTCAGA |
|


|
|
Erra(NR)/HepG2-Erra-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 2 |
| Score: | 0.72
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -AGAGGGCA-
CAAAGGTCAG |
|


|
|
MA0597.1_THAP1/Jaspar
| Match Rank: | 3 |
| Score: | 0.71
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | AGAGGGCA-
TNNGGGCAG |
|


|
|
PB0118.1_Esrra_2/Jaspar
| Match Rank: | 4 |
| Score: | 0.69
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----AGAGGGCA-----
GGCGAGGGGTCAAGGGC |
|


|
|
MA0065.2_PPARG::RXRA/Jaspar
| Match Rank: | 5 |
| Score: | 0.68
| | Offset: | -7
| | Orientation: | forward strand |
| Alignment: | -------AGAGGGCA
GTAGGGCAAAGGTCA |
|


|
|
PB0049.1_Nr2f2_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.68
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----AGAGGGCA----
TCTCAAAGGTCACGAG |
|


|
|
RXR(NR/DR1)/3T3L1-RXR-ChIP-Seq(GSE13511)/Homer
| Match Rank: | 7 |
| Score: | 0.67
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------AGAGGGCA
TAGGGCAAAGGTCA |
|


|
|
PB0053.1_Rara_1/Jaspar
| Match Rank: | 8 |
| Score: | 0.67
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----AGAGGGCA----
TCTCAAAGGTCACCTG |
|


|
|
PB0133.1_Hic1_2/Jaspar
| Match Rank: | 9 |
| Score: | 0.66
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---AGAGGGCA-----
NNNNTTGGGCACNNCN |
|

|
|
CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer
| Match Rank: | 10 |
| Score: | 0.66
| | Offset: | -8
| | Orientation: | reverse strand |
| Alignment: | --------AGAGGGCA----
TGGCCACCAGGTGGCACTNT |
|


|
|





















