| p-value: | 1e-11 |
| log p-value: | -2.587e+01 |
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 23.0 |
| Percentage of Target Sequences with motif | 2.73% |
| Number of Background Sequences with motif | 3.3 |
| Percentage of Background Sequences with motif | 0.47% |
| Average Position of motif in Targets | 93.3 +/- 54.1bp |
| Average Position of motif in Background | 110.3 +/- 56.9bp |
| Strand Bias (log2 ratio + to - strand density) | 1.1 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
MA0117.1_Mafb/Jaspar
| Match Rank: | 1 |
| Score: | 0.67
| | Offset: | 4
| | Orientation: | forward strand |
| Alignment: | CCACGCTGACAC
----GCTGACGC |
|


|
|
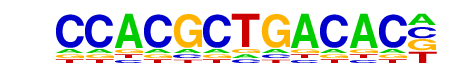
MafA(bZIP)/Islet-MafA-ChIP-Seq(GSE30298)/Homer
| Match Rank: | 2 |
| Score: | 0.65
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | CCACGCTGACAC-
---TGCTGACTCA |
|

|
|
MA0006.1_Arnt::Ahr/Jaspar
| Match Rank: | 3 |
| Score: | 0.61
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CCACGCTGACAC
-CACGCA----- |
|


|
|
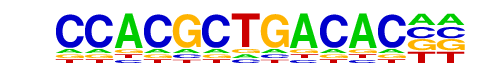
Tbx5(T-box)/HL1-Tbx5.biotin-ChIP-Seq(GSE21529)/Homer
| Match Rank: | 4 |
| Score: | 0.57
| | Offset: | 6
| | Orientation: | reverse strand |
| Alignment: | CCACGCTGACAC--
------TGACACCT |
|

|
|
MA0258.2_ESR2/Jaspar
| Match Rank: | 5 |
| Score: | 0.56
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---CCACGCTGACAC
AGGNCANNGTGACCT |
|


|
|
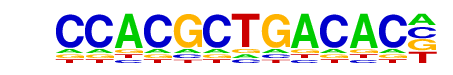
Tbx20(T-box)/Heart-Tbx20-ChIP-Seq(GSE29636)/Homer
| Match Rank: | 6 |
| Score: | 0.56
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CCACGCTGACAC-
-GGTGYTGACAGS |
|

|
|
MA0498.1_Meis1/Jaspar
| Match Rank: | 7 |
| Score: | 0.55
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | CCACGCTGACAC------
---AGCTGTCACTCACCT |
|


|
|
ERE(NR/IR3)/MCF7-ERa-ChIP-Seq(Unpublished)/Homer
| Match Rank: | 8 |
| Score: | 0.54
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CCACGCTGACAC-
GGTCANNGTGACCTN |
|


|
|
MA0130.1_ZNF354C/Jaspar
| Match Rank: | 9 |
| Score: | 0.54
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CCACGCTGACAC
ATCCAC-------- |
|


|
|
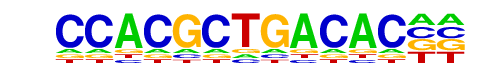
Tbet(T-box)/CD8-Tbet-ChIP-Seq(GSE33802)/Homer
| Match Rank: | 10 |
| Score: | 0.54
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | CCACGCTGACAC--
----KTTCACACCT |
|

|
|





















