
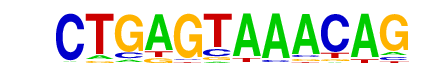






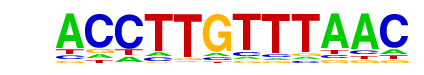


| p-value: | 1e-142 |
| log p-value: | -3.275e+02 |
| Information Content per bp: | 1.688 |
| Number of Target Sequences with motif | 630.0 |
| Percentage of Target Sequences with motif | 19.85% |
| Number of Background Sequences with motif | 177.2 |
| Percentage of Background Sequences with motif | 6.33% |
| Average Position of motif in Targets | 99.7 +/- 52.3bp |
| Average Position of motif in Background | 97.8 +/- 58.5bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.16 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.901 |  | 1e-98 | -227.067963 | 17.01% | 6.18% | motif file (matrix) |
| 2 | 0.763 |  | 1e-58 | -134.267032 | 4.06% | 0.65% | motif file (matrix) |
| 3 | 0.755 |  | 1e-56 | -129.153333 | 1.76% | 0.09% | motif file (matrix) |
| 4 | 0.700 |  | 1e-49 | -113.487686 | 13.71% | 6.38% | motif file (matrix) |
| 5 | 0.750 |  | 1e-39 | -91.692264 | 13.26% | 6.68% | motif file (matrix) |
| 6 | 0.840 |  | 1e-36 | -83.450804 | 1.80% | 0.19% | motif file (matrix) |
| 7 | 0.613 |  | 1e-33 | -76.006681 | 3.72% | 0.99% | motif file (matrix) |
| 8 | 0.738 |  | 1e-32 | -74.242894 | 1.20% | 0.07% | motif file (matrix) |
| 9 | 0.618 |  | 1e-23 | -55.017650 | 0.98% | 0.09% | motif file (matrix) |