
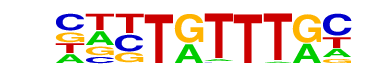







| p-value: | 1e-112 |
| log p-value: | -2.596e+02 |
| Information Content per bp: | 1.558 |
| Number of Target Sequences with motif | 904.0 |
| Percentage of Target Sequences with motif | 28.48% |
| Number of Background Sequences with motif | 6138.6 |
| Percentage of Background Sequences with motif | 13.19% |
| Average Position of motif in Targets | 100.5 +/- 52.5bp |
| Average Position of motif in Background | 100.5 +/- 58.5bp |
| Strand Bias (log2 ratio + to - strand density) | -0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.19 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.918 |  | 1e-105 | -242.229895 | 30.78% | 15.33% | motif file (matrix) |
| 2 | 0.883 |  | 1e-84 | -195.359986 | 28.99% | 15.26% | motif file (matrix) |
| 3 | 0.906 |  | 1e-83 | -193.073148 | 12.26% | 3.94% | motif file (matrix) |
| 4 | 0.900 |  | 1e-67 | -154.473385 | 24.98% | 13.44% | motif file (matrix) |
| 5 | 0.625 |  | 1e-43 | -99.084546 | 23.94% | 14.62% | motif file (matrix) |
| 6 | 0.742 |  | 1e-38 | -89.171454 | 15.78% | 8.58% | motif file (matrix) |
| 7 | 0.694 |  | 1e-23 | -54.798664 | 10.87% | 6.10% | motif file (matrix) |