| p-value: | 1e-27 |
| log p-value: | -6.239e+01 |
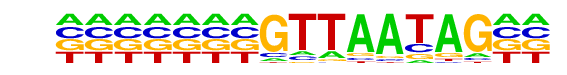
| Information Content per bp: | 1.597 |
| Number of Target Sequences with motif | 33.0 |
| Percentage of Target Sequences with motif | 18.64% |
| Number of Background Sequences with motif | 2.9 |
| Percentage of Background Sequences with motif | 1.87% |
| Average Position of motif in Targets | 110.6 +/- 45.7bp |
| Average Position of motif in Background | 88.9 +/- 49.4bp |
| Strand Bias (log2 ratio + to - strand density) | -0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.10 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PB0109.1_Bbx_2/Jaspar
| Match Rank: | 1 |
| Score: | 0.81
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----GTTAATAG----
TGATTGTTAACAGTTGG |
|


|
|
PB0135.1_Hoxa3_2/Jaspar
| Match Rank: | 2 |
| Score: | 0.73
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -GTTAATAG-----
CCTTAATNGNTTTT |
|


|
|
MA0063.1_Nkx2-5/Jaspar
| Match Rank: | 3 |
| Score: | 0.73
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | GTTAATAG
-TTAATTG |
|


|
|
Nanog(Homeobox)/mES-Nanog-ChIP-Seq(GSE11724)/Homer
| Match Rank: | 4 |
| Score: | 0.72
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GTTAATAG--
GTTAATGGCC |
|


|
|
Nkx6.1(Homeobox)/Islet-Nkx6.1-ChIP-Seq(GSE40975)/Homer
| Match Rank: | 5 |
| Score: | 0.70
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GTTAATAG
GKTAATGR |
|


|
|
Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan et al.)/Homer
| Match Rank: | 6 |
| Score: | 0.69
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GTTAATAG
GTAAACAG |
|


|
|
Foxa2(Forkhead)/Liver-Foxa2-ChIP-Seq(GSE25694)/Homer
| Match Rank: | 7 |
| Score: | 0.69
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---GTTAATAG-
TATGTAAACANG |
|


|
|
Isl1(Homeobox)/Neuron-Isl1-ChIP-Seq(GSE31456)/Homer
| Match Rank: | 8 |
| Score: | 0.68
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | GTTAATAG-
-CTAATKGV |
|


|
|
Unknown(Homeobox)/Limb-p300-ChIP-Seq/Homer
| Match Rank: | 9 |
| Score: | 0.68
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GTTAATAG--
TTTAATTGCN |
|


|
|
PH0168.1_Hnf1b/Jaspar
| Match Rank: | 10 |
| Score: | 0.67
| | Offset: | -7
| | Orientation: | reverse strand |
| Alignment: | -------GTTAATAG--
ANNNCTAGTTAACNGNN |
|

|
|





















