
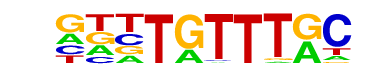












| p-value: | 1e-1214 |
| log p-value: | -2.798e+03 |
| Information Content per bp: | 1.656 |
| Number of Target Sequences with motif | 6389.0 |
| Percentage of Target Sequences with motif | 15.87% |
| Number of Background Sequences with motif | 2221.4 |
| Percentage of Background Sequences with motif | 5.55% |
| Average Position of motif in Targets | 99.4 +/- 52.2bp |
| Average Position of motif in Background | 100.1 +/- 59.3bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.14 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.934 |  | 1e-1166 | -2685.389483 | 24.42% | 11.35% | motif file (matrix) |
| 2 | 0.933 |  | 1e-1086 | -2501.530247 | 24.98% | 12.12% | motif file (matrix) |
| 3 | 0.665 |  | 1e-715 | -1646.382543 | 33.46% | 21.12% | motif file (matrix) |
| 4 | 0.777 |  | 1e-644 | -1483.210248 | 13.82% | 6.28% | motif file (matrix) |
| 5 | 0.706 |  | 1e-270 | -623.371515 | 18.20% | 12.12% | motif file (matrix) |
| 6 | 0.615 |  | 1e-253 | -582.803065 | 13.25% | 8.22% | motif file (matrix) |
| 7 | 0.699 |  | 1e-166 | -383.219677 | 2.37% | 0.84% | motif file (matrix) |
| 8 | 0.711 |  | 1e-141 | -325.109152 | 3.89% | 1.92% | motif file (matrix) |
| 9 | 0.626 |  | 1e-128 | -295.509724 | 3.68% | 1.85% | motif file (matrix) |
| 10 | 0.723 |  | 1e-92 | -213.110497 | 0.90% | 0.25% | motif file (matrix) |
| 11 | 0.613 |  | 1e-85 | -196.645070 | 0.34% | 0.03% | motif file (matrix) |
| 12 | 0.702 |  | 1e-25 | -57.569210 | 0.17% | 0.04% | motif file (matrix) |