| p-value: | 1e-152 |
| log p-value: | -3.517e+02 |
| Information Content per bp: | 1.835 |
| Number of Target Sequences with motif | 5800.0 |
| Percentage of Target Sequences with motif | 22.47% |
| Number of Background Sequences with motif | 2852.4 |
| Percentage of Background Sequences with motif | 16.15% |
| Average Position of motif in Targets | 99.9 +/- 54.9bp |
| Average Position of motif in Background | 101.6 +/- 75.7bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.15 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
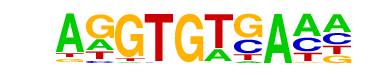
Tbet(T-box)/CD8-Tbet-ChIP-Seq(GSE33802)/Homer
| Match Rank: | 1 |
| Score: | 0.94
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AGGTGTGA--
AGGTGTGAAM |
|

|
|
Tbx5(T-box)/HL1-Tbx5.biotin-ChIP-Seq(GSE21529)/Homer
| Match Rank: | 2 |
| Score: | 0.91
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AGGTGTGA
AGGTGTCA |
|


|
|
PB0013.1_Eomes_1/Jaspar
| Match Rank: | 3 |
| Score: | 0.90
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----AGGTGTGA-----
GAAAAGGTGTGAAAATT |
|


|
|
MA0009.1_T/Jaspar
| Match Rank: | 4 |
| Score: | 0.89
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --AGGTGTGA-
CTAGGTGTGAA |
|


|
|
Eomes(T-box)/H9-Eomes-ChIP-Seq(GSE26097)/Homer
| Match Rank: | 5 |
| Score: | 0.86
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | AGGTGTGA--
AGGTGTTAAT |
|


|
|
MA0103.2_ZEB1/Jaspar
| Match Rank: | 6 |
| Score: | 0.77
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -AGGTGTGA
CAGGTGAGG |
|


|
|
E2A-nearPU.1(HLH)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 7 |
| Score: | 0.76
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---AGGTGTGA
NNCAGGTGNN- |
|


|
|
PH0164.1_Six4/Jaspar
| Match Rank: | 8 |
| Score: | 0.75
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----AGGTGTGA-----
TNNNNGGTGTCATNTNT |
|


|
|
PB0089.1_Tcfe2a_1/Jaspar
| Match Rank: | 9 |
| Score: | 0.71
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------AGGTGTGA---
ATCCACAGGTGCGAAAA |
|


|
|
PB0117.1_Eomes_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.70
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----AGGTGTGA----
GCGGAGGTGTCGCCTC |
|


|
|





















