








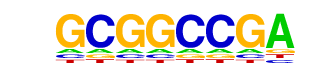




| p-value: | 1e-273 |
| log p-value: | -6.298e+02 |
| Information Content per bp: | 1.734 |
| Number of Target Sequences with motif | 167.0 |
| Percentage of Target Sequences with motif | 0.65% |
| Number of Background Sequences with motif | 1.9 |
| Percentage of Background Sequences with motif | 0.01% |
| Average Position of motif in Targets | 106.3 +/- 53.7bp |
| Average Position of motif in Background | 69.0 +/- 0.0bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.04 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.657 |  | 1e-250 | -577.950476 | 0.60% | 0.00% | motif file (matrix) |
| 2 | 0.785 |  | 1e-147 | -339.174635 | 0.53% | 0.02% | motif file (matrix) |
| 3 | 0.621 |  | 1e-131 | -302.209174 | 0.36% | 0.01% | motif file (matrix) |
| 4 | 0.600 |  | 1e-95 | -221.040766 | 0.29% | 0.00% | motif file (matrix) |
| 5 | 0.709 |  | 1e-86 | -198.039017 | 0.57% | 0.07% | motif file (matrix) |
| 6 | 0.776 |  | 1e-76 | -175.344898 | 0.49% | 0.06% | motif file (matrix) |
| 7 | 0.686 |  | 1e-71 | -164.729226 | 0.28% | 0.01% | motif file (matrix) |
| 8 | 0.719 |  | 1e-58 | -134.664633 | 0.38% | 0.04% | motif file (matrix) |
| 9 | 0.704 |  | 1e-36 | -84.143716 | 0.59% | 0.18% | motif file (matrix) |
| 10 | 0.634 |  | 1e-28 | -64.959210 | 0.53% | 0.17% | motif file (matrix) |
| 11 | 0.664 |  | 1e-27 | -64.180843 | 0.43% | 0.12% | motif file (matrix) |
| 12 | 0.635 |  | 1e-6 | -15.882964 | 3.06% | 2.55% | motif file (matrix) |