













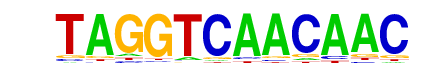



| p-value: | 1e-956 |
| log p-value: | -2.202e+03 |
| Information Content per bp: | 1.608 |
| Number of Target Sequences with motif | 6066.0 |
| Percentage of Target Sequences with motif | 23.50% |
| Number of Background Sequences with motif | 2421.2 |
| Percentage of Background Sequences with motif | 9.45% |
| Average Position of motif in Targets | 99.6 +/- 52.5bp |
| Average Position of motif in Background | 99.4 +/- 60.1bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.20 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.968 |  | 1e-828 | -1908.538505 | 21.45% | 8.81% | motif file (matrix) |
| 2 | 0.941 |  | 1e-767 | -1766.890041 | 25.16% | 11.75% | motif file (matrix) |
| 3 | 0.807 |  | 1e-451 | -1039.334995 | 33.61% | 21.34% | motif file (matrix) |
| 4 | 0.837 |  | 1e-365 | -841.387072 | 14.42% | 7.06% | motif file (matrix) |
| 5 | 0.612 |  | 1e-347 | -799.644869 | 30.56% | 20.07% | motif file (matrix) |
| 6 | 0.769 |  | 1e-325 | -750.024826 | 29.44% | 19.40% | motif file (matrix) |
| 7 | 0.751 |  | 1e-246 | -566.932251 | 6.96% | 2.86% | motif file (matrix) |
| 8 | 0.736 |  | 1e-242 | -558.568161 | 14.60% | 8.33% | motif file (matrix) |
| 9 | 0.669 |  | 1e-120 | -277.988811 | 5.59% | 2.86% | motif file (matrix) |
| 10 | 0.717 |  | 1e-96 | -222.085871 | 2.03% | 0.70% | motif file (matrix) |
| 11 | 0.621 |  | 1e-53 | -122.539920 | 0.35% | 0.04% | motif file (matrix) |
| 12 | 0.769 |  | 1e-38 | -89.256406 | 0.28% | 0.04% | motif file (matrix) |
| 13 | 0.628 |  | 1e-22 | -52.482496 | 0.29% | 0.07% | motif file (matrix) |
| 14 | 0.623 |  | 1e-16 | -38.652375 | 0.14% | 0.03% | motif file (matrix) |
| 15 | 0.621 |  | 1e-14 | -32.850096 | 0.12% | 0.02% | motif file (matrix) |
| 16 | 0.615 |  | 1e-13 | -30.972912 | 0.15% | 0.04% | motif file (matrix) |