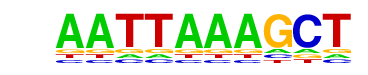
| p-value: | 1e-11 |
| log p-value: | -2.748e+01 |
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 28.0 |
| Percentage of Target Sequences with motif | 0.11% |
| Number of Background Sequences with motif | 5.6 |
| Percentage of Background Sequences with motif | 0.02% |
| Average Position of motif in Targets | 96.4 +/- 50.7bp |
| Average Position of motif in Background | 47.0 +/- 0.0bp |
| Strand Bias (log2 ratio + to - strand density) | 0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
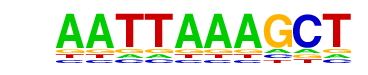
MA0151.1_ARID3A/Jaspar
| Match Rank: | 1 |
| Score: | 0.74
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | AATTAAAGCT
-ATTAAA--- |
|

|
|
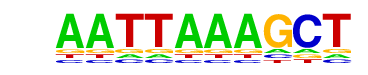
MA0075.1_Prrx2/Jaspar
| Match Rank: | 2 |
| Score: | 0.71
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AATTAAAGCT
AATTA----- |
|

|
|
PH0041.1_Hmx1/Jaspar
| Match Rank: | 3 |
| Score: | 0.70
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------AATTAAAGCT-
ACAAGCAATTAATGAAT |
|


|
|
PH0156.1_Rax/Jaspar
| Match Rank: | 4 |
| Score: | 0.70
| | Offset: | -7
| | Orientation: | reverse strand |
| Alignment: | -------AATTAAAGCT
NNGCGCTAATTAGTGCA |
|


|
|
PH0011.1_Alx1_2/Jaspar
| Match Rank: | 5 |
| Score: | 0.70
| | Offset: | -7
| | Orientation: | reverse strand |
| Alignment: | -------AATTAAAGCT
NNNAATTAATTAANGNG |
|


|
|
PH0002.1_Alx4/Jaspar
| Match Rank: | 6 |
| Score: | 0.69
| | Offset: | -7
| | Orientation: | reverse strand |
| Alignment: | -------AATTAAAGCT
GNNNATTAATTAATGCG |
|


|
|
PH0054.1_Hoxa7_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.69
| | Offset: | -7
| | Orientation: | reverse strand |
| Alignment: | -------AATTAAAGCT
GNNNATTAATTAANNCG |
|


|
|
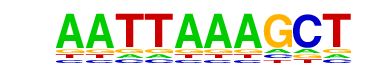
MA0158.1_HOXA5/Jaspar
| Match Rank: | 8 |
| Score: | 0.69
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | AATTAAAGCT
AATTAGTG-- |
|

|
|
Unknown(Homeobox)/Limb-p300-ChIP-Seq/Homer
| Match Rank: | 9 |
| Score: | 0.69
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---AATTAAAGCT
NGCAATTAAA--- |
|


|
|
PH0010.1_Alx1_1/Jaspar
| Match Rank: | 10 |
| Score: | 0.69
| | Offset: | -7
| | Orientation: | reverse strand |
| Alignment: | -------AATTAAAGCT
GNNNATTAATTAATNCG |
|


|
|