| p-value: | 1e-21 |
| log p-value: | -4.985e+01 |
| Information Content per bp: | 1.676 |
| Number of Target Sequences with motif | 30.0 |
| Percentage of Target Sequences with motif | 0.49% |
| Number of Background Sequences with motif | 2.9 |
| Percentage of Background Sequences with motif | 0.06% |
| Average Position of motif in Targets | 96.8 +/- 56.4bp |
| Average Position of motif in Background | 92.5 +/- 50.7bp |
| Strand Bias (log2 ratio + to - strand density) | -0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.04 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
AR-halfsite(NR)/LNCaP-AR-ChIP-Seq(GSE27824)/Homer
| Match Rank: | 1 |
| Score: | 0.64
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TGTCCGAGCG
CTGTTCCTGG- |
|


|
|
PB0140.1_Irf6_2/Jaspar
| Match Rank: | 2 |
| Score: | 0.56
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TGTCCGAGCG----
NNNACCGAGAGTNNN |
|


|
|
POL010.1_DCE_S_III/Jaspar
| Match Rank: | 3 |
| Score: | 0.55
| | Offset: | 5
| | Orientation: | forward strand |
| Alignment: | TGTCCGAGCG
-----CAGCC |
|


|
|
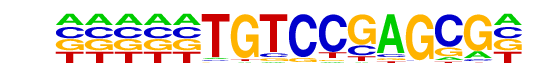
PB0133.1_Hic1_2/Jaspar
| Match Rank: | 4 |
| Score: | 0.55
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----TGTCCGAGCG-
GGGTGTGCCCAAAAGG |
|

|
|
Nrf2(bZIP)/Lymphoblast-Nrf2-ChIP-Seq(GSE37589)/Homer
| Match Rank: | 5 |
| Score: | 0.54
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TGTCCGAGCG-
ATGACTCAGCAD |
|


|
|
PB0153.1_Nr2f2_2/Jaspar
| Match Rank: | 6 |
| Score: | 0.54
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TGTCCGAGCG--
NNNNTGACCCGGCGCG |
|


|
|
p53(p53)/mES-cMyc-ChIP-Seq(GSE11431)/Homer
| Match Rank: | 7 |
| Score: | 0.54
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TGTCCGAGCG---
ATGCCCGGGCATGT |
|


|
|
PB0086.1_Tcfap2b_1/Jaspar
| Match Rank: | 8 |
| Score: | 0.54
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TGTCCGAGCG---
NTGCCCTAGGGCAA |
|


|
|
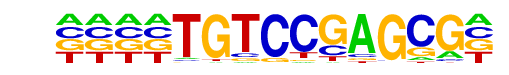
MA0591.1_Bach1::Mafk/Jaspar
| Match Rank: | 9 |
| Score: | 0.54
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----TGTCCGAGCG-
AGGATGACTCAGCAC |
|

|
|
PB0138.1_Irf4_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.53
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TGTCCGAGCG----
GNNACCGAGAATNNN |
|


|
|





















