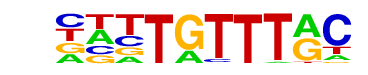
| p-value: | 1e-151 |
| log p-value: | -3.493e+02 |
| Information Content per bp: | 1.647 |
| Number of Target Sequences with motif | 1709.0 |
| Percentage of Target Sequences with motif | 27.91% |
| Number of Background Sequences with motif | 6468.2 |
| Percentage of Background Sequences with motif | 14.82% |
| Average Position of motif in Targets | 101.2 +/- 51.7bp |
| Average Position of motif in Background | 98.5 +/- 61.1bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.25 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer
| Match Rank: | 1 |
| Score: | 0.92
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --GYAAACAANN
NDGTAAACARRN |
|


|
|
MA0593.1_FOXP2/Jaspar
| Match Rank: | 2 |
| Score: | 0.91
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --GYAAACAANN
AAGTAAACAAA- |
|


|
|
MA0031.1_FOXD1/Jaspar
| Match Rank: | 3 |
| Score: | 0.91
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GYAAACAANN
GTAAACAT-- |
|


|
|
MF0005.1_Forkhead_class/Jaspar
| Match Rank: | 4 |
| Score: | 0.90
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --GYAAACAANN
AAATAAACA--- |
|


|
|
Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan et al.)/Homer
| Match Rank: | 5 |
| Score: | 0.90
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GYAAACAANN
GTAAACAG-- |
|


|
|
MA0157.1_FOXO3/Jaspar
| Match Rank: | 6 |
| Score: | 0.90
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GYAAACAANN
TGTAAACA--- |
|


|
|
PB0017.1_Foxj3_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.89
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----GYAAACAANN--
AAAAAGTAAACAAACAC |
|


|
|
PB0016.1_Foxj1_1/Jaspar
| Match Rank: | 8 |
| Score: | 0.89
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---GYAAACAANN---
AAAGTAAACAAAAATT |
|


|
|
MA0030.1_FOXF2/Jaspar
| Match Rank: | 9 |
| Score: | 0.89
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----GYAAACAANN
CAAACGTAAACAAT- |
|


|
|
FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer
| Match Rank: | 10 |
| Score: | 0.88
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---GYAAACAANN
AAAGTAAACA--- |
|


|
|