







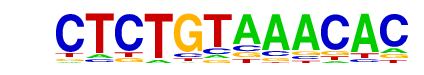




| p-value: | 1e-151 |
| log p-value: | -3.493e+02 |
| Information Content per bp: | 1.647 |
| Number of Target Sequences with motif | 1709.0 |
| Percentage of Target Sequences with motif | 27.91% |
| Number of Background Sequences with motif | 6468.2 |
| Percentage of Background Sequences with motif | 14.82% |
| Average Position of motif in Targets | 101.2 +/- 51.7bp |
| Average Position of motif in Background | 98.5 +/- 61.1bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.25 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
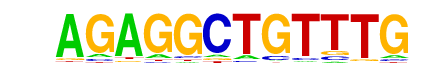
| 1 | 0.989 |  | 1e-137 | -316.378849 | 36.91% | 22.73% | motif file (matrix) |
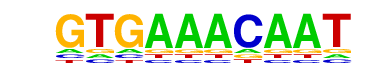
| 2 | 0.934 |  | 1e-117 | -270.536867 | 26.93% | 15.39% | motif file (matrix) |
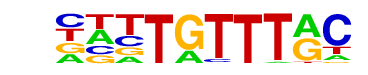
| 3 | 0.688 |  | 1e-97 | -224.316444 | 14.50% | 6.81% | motif file (matrix) |
| 4 | 0.610 |  | 1e-71 | -164.575751 | 23.37% | 14.69% | motif file (matrix) |
| 5 | 0.643 |  | 1e-68 | -157.536456 | 24.69% | 15.96% | motif file (matrix) |
| 6 | 0.780 |  | 1e-43 | -99.797836 | 9.85% | 5.40% | motif file (matrix) |
| 7 | 0.718 |  | 1e-38 | -89.112623 | 10.57% | 6.16% | motif file (matrix) |
| 8 | 0.680 |  | 1e-38 | -87.818140 | 4.85% | 2.08% | motif file (matrix) |
| 9 | 0.644 |  | 1e-33 | -76.710617 | 9.80% | 5.84% | motif file (matrix) |
| 10 | 0.707 |  | 1e-28 | -65.504935 | 37.91% | 31.18% | motif file (matrix) |
| 11 | 0.604 |  | 1e-18 | -43.092973 | 3.89% | 2.06% | motif file (matrix) |
| 12 | 0.618 |  | 1e-11 | -25.583197 | 0.21% | 0.02% | motif file (matrix) |
| 13 | 0.611 |  | 1e-9 | -21.530594 | 1.01% | 0.41% | motif file (matrix) |
| 14 | 0.617 |  | 1e-4 | -9.306203 | 0.10% | 0.01% | motif file (matrix) |