








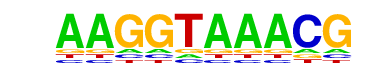


| p-value: | 1e-186 |
| log p-value: | -4.299e+02 |
| Information Content per bp: | 1.583 |
| Number of Target Sequences with motif | 1814.0 |
| Percentage of Target Sequences with motif | 31.48% |
| Number of Background Sequences with motif | 7004.2 |
| Percentage of Background Sequences with motif | 15.96% |
| Average Position of motif in Targets | 98.8 +/- 52.2bp |
| Average Position of motif in Background | 100.3 +/- 59.5bp |
| Strand Bias (log2 ratio + to - strand density) | 0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.26 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.949 |  | 1e-163 | -376.692743 | 31.18% | 16.57% | motif file (matrix) |
| 2 | 0.963 |  | 1e-159 | -367.669967 | 23.65% | 11.07% | motif file (matrix) |
| 3 | 0.839 |  | 1e-113 | -260.692385 | 31.55% | 19.02% | motif file (matrix) |
| 4 | 0.844 |  | 1e-98 | -226.136768 | 20.02% | 10.57% | motif file (matrix) |
| 5 | 0.779 |  | 1e-50 | -117.237159 | 15.25% | 9.05% | motif file (matrix) |
| 6 | 0.747 |  | 1e-33 | -76.467077 | 6.78% | 3.49% | motif file (matrix) |
| 7 | 0.752 |  | 1e-32 | -73.753511 | 5.74% | 2.80% | motif file (matrix) |
| 8 | 0.666 |  | 1e-18 | -43.182411 | 1.04% | 0.25% | motif file (matrix) |
| 9 | 0.650 |  | 1e-8 | -19.794985 | 2.72% | 1.65% | motif file (matrix) |
| 10 | 0.625 |  | 1e-6 | -14.828147 | 0.10% | 0.01% | motif file (matrix) |