





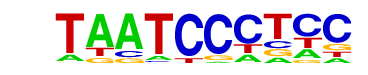






| p-value: | 1e-135 |
| log p-value: | -3.126e+02 |
| Information Content per bp: | 1.600 |
| Number of Target Sequences with motif | 1998.0 |
| Percentage of Target Sequences with motif | 34.67% |
| Number of Background Sequences with motif | 9014.7 |
| Percentage of Background Sequences with motif | 20.54% |
| Average Position of motif in Targets | 99.1 +/- 53.2bp |
| Average Position of motif in Background | 100.3 +/- 59.3bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.30 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.895 |  | 1e-128 | -295.272894 | 24.02% | 12.41% | motif file (matrix) |
| 2 | 0.820 |  | 1e-69 | -159.485529 | 27.47% | 18.01% | motif file (matrix) |
| 3 | 0.819 |  | 1e-61 | -141.435032 | 12.55% | 6.51% | motif file (matrix) |
| 4 | 0.630 |  | 1e-60 | -140.446420 | 32.55% | 23.00% | motif file (matrix) |
| 5 | 0.702 |  | 1e-57 | -131.909912 | 21.00% | 13.32% | motif file (matrix) |
| 6 | 0.756 |  | 1e-45 | -104.384687 | 6.52% | 2.88% | motif file (matrix) |
| 7 | 0.645 |  | 1e-41 | -94.673179 | 12.46% | 7.38% | motif file (matrix) |
| 8 | 0.710 |  | 1e-37 | -85.657047 | 5.97% | 2.78% | motif file (matrix) |
| 9 | 0.630 |  | 1e-33 | -78.254632 | 8.95% | 5.06% | motif file (matrix) |
| 10 | 0.637 |  | 1e-24 | -56.952132 | 5.52% | 2.94% | motif file (matrix) |
| 11 | 0.608 |  | 1e-13 | -30.750629 | 1.09% | 0.36% | motif file (matrix) |