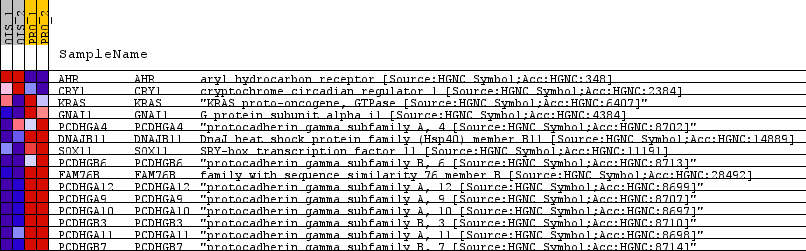
| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | | 1 | AHR | aryl hydrocarbon receptor [Source:HGNC Symbol;Acc:HGNC:348] | 96 | 0.988 | -0.0451 | No |
| 2 | CRY1 | cryptochrome circadian regulator 1 [Source:HGNC Symbol;Acc:HGNC:2384] | 269 | 0.182 | -0.3107 | No |
| 3 | KRAS | "KRAS proto-oncogene, GTPase [Source:HGNC Symbol;Acc:HGNC:6407]" | 369 | -0.041 | -0.4709 | No |
| 4 | GNAI1 | G protein subunit alpha i1 [Source:HGNC Symbol;Acc:HGNC:4384] | 397 | -0.171 | -0.4960 | No |
| 5 | PCDHGA4 | "protocadherin gamma subfamily A, 4 [Source:HGNC Symbol;Acc:HGNC:8702]" | 435 | -0.300 | -0.5228 | Yes |
| 6 | DNAJB11 | DnaJ heat shock protein family (Hsp40) member B11 [Source:HGNC Symbol;Acc:HGNC:14889] | 446 | -0.348 | -0.4990 | Yes |
| 7 | SOX11 | SRY-box transcription factor 11 [Source:HGNC Symbol;Acc:HGNC:11191] | 449 | -0.355 | -0.4610 | Yes |
| 8 | PCDHGB6 | "protocadherin gamma subfamily B, 6 [Source:HGNC Symbol;Acc:HGNC:8713]" | 462 | -0.404 | -0.4340 | Yes |
| 9 | FAM76B | family with sequence similarity 76 member B [Source:HGNC Symbol;Acc:HGNC:28492] | 536 | -0.760 | -0.4674 | Yes |
| 10 | PCDHGA12 | "protocadherin gamma subfamily A, 12 [Source:HGNC Symbol;Acc:HGNC:8699]" | 541 | -0.771 | -0.3844 | Yes |
| 11 | PCDHGA9 | "protocadherin gamma subfamily A, 9 [Source:HGNC Symbol;Acc:HGNC:8707]" | 543 | -0.778 | -0.2956 | Yes |
| 12 | PCDHGA10 | "protocadherin gamma subfamily A, 10 [Source:HGNC Symbol;Acc:HGNC:8697]" | 547 | -0.791 | -0.2087 | Yes |
| 13 | PCDHGB3 | "protocadherin gamma subfamily B, 3 [Source:HGNC Symbol;Acc:HGNC:8710]" | 552 | -0.817 | -0.1204 | Yes |
| 14 | PCDHGA11 | "protocadherin gamma subfamily A, 11 [Source:HGNC Symbol;Acc:HGNC:8698]" | 556 | -0.872 | -0.0240 | Yes |
| 15 | PCDHGB7 | "protocadherin gamma subfamily B, 7 [Source:HGNC Symbol;Acc:HGNC:8714]" | 570 | -1.023 | 0.0733 | Yes |
Table: GSEA details [plain text format]