| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | | 1 | RGS5 | regulator of G protein signaling 5 [Source:HGNC Symbol;Acc:HGNC:10001] | 105 | 0.916 | -0.1332 | No |
| 2 | DTX4 | deltex E3 ubiquitin ligase 4 [Source:HGNC Symbol;Acc:HGNC:29151] | 112 | 0.881 | -0.0982 | No |
| 3 | TSSC4 | tumor suppressing subtransferable candidate 4 [Source:HGNC Symbol;Acc:HGNC:12386] | 114 | 0.839 | -0.0567 | No |
| 4 | SIK3 | SIK family kinase 3 [Source:HGNC Symbol;Acc:HGNC:29165] | 169 | 0.471 | -0.1253 | No |
| 5 | MAP3K5 | mitogen-activated protein kinase kinase kinase 5 [Source:HGNC Symbol;Acc:HGNC:6857] | 178 | 0.448 | -0.1160 | No |
| 6 | NCOR2 | nuclear receptor corepressor 2 [Source:HGNC Symbol;Acc:HGNC:7673] | 188 | 0.405 | -0.1106 | No |
| 7 | TIMM13 | translocase of inner mitochondrial membrane 13 [Source:HGNC Symbol;Acc:HGNC:11816] | 194 | 0.400 | -0.0986 | No |
| 8 | RPL37 | ribosomal protein L37 [Source:HGNC Symbol;Acc:HGNC:10347] | 212 | 0.340 | -0.1103 | No |
| 9 | MIEF1 | mitochondrial elongation factor 1 [Source:HGNC Symbol;Acc:HGNC:25979] | 219 | 0.320 | -0.1041 | No |
| 10 | CRLS1 | cardiolipin synthase 1 [Source:HGNC Symbol;Acc:HGNC:16148] | 234 | 0.279 | -0.1138 | No |
| 11 | ANAPC13 | anaphase promoting complex subunit 13 [Source:HGNC Symbol;Acc:HGNC:24540] | 245 | 0.251 | -0.1181 | No |
| 12 | NXT1 | nuclear transport factor 2 like export factor 1 [Source:HGNC Symbol;Acc:HGNC:15913] | 256 | 0.215 | -0.1242 | No |
| 13 | TMX2 | thioredoxin related transmembrane protein 2 [Source:HGNC Symbol;Acc:HGNC:30739] | 264 | 0.198 | -0.1260 | No |
| 14 | GZF1 | GDNF inducible zinc finger protein 1 [Source:HGNC Symbol;Acc:HGNC:15808] | 307 | 0.101 | -0.1929 | No |
| 15 | MED19 | mediator complex subunit 19 [Source:HGNC Symbol;Acc:HGNC:29600] | 314 | 0.079 | -0.1992 | No |
| 16 | COPS2 | COP9 signalosome subunit 2 [Source:HGNC Symbol;Acc:HGNC:30747] | 349 | 0.001 | -0.2575 | No |
| 17 | SETD4 | SET domain containing 4 [Source:HGNC Symbol;Acc:HGNC:1258] | 362 | -0.037 | -0.2763 | No |
| 18 | OSTC | oligosaccharyltransferase complex non-catalytic subunit [Source:HGNC Symbol;Acc:HGNC:24448] | 374 | -0.059 | -0.2921 | No |
| 19 | TOE1 | "target of EGR1, exonuclease [Source:HGNC Symbol;Acc:HGNC:15954]" | 398 | -0.176 | -0.3226 | No |
| 20 | GALK2 | galactokinase 2 [Source:HGNC Symbol;Acc:HGNC:4119] | 410 | -0.238 | -0.3293 | No |
| 21 | DCP2 | decapping mRNA 2 [Source:HGNC Symbol;Acc:HGNC:24452] | 455 | -0.375 | -0.3855 | Yes |
| 22 | RNF32 | ring finger protein 32 [Source:HGNC Symbol;Acc:HGNC:17118] | 461 | -0.399 | -0.3736 | Yes |
| 23 | RAD52 | "RAD52 homolog, DNA repair protein [Source:HGNC Symbol;Acc:HGNC:9824]" | 468 | -0.419 | -0.3623 | Yes |
| 24 | SLC38A1 | solute carrier family 38 member 1 [Source:HGNC Symbol;Acc:HGNC:13447] | 489 | -0.511 | -0.3704 | Yes |
| 25 | MGST3 | microsomal glutathione S-transferase 3 [Source:HGNC Symbol;Acc:HGNC:7064] | 495 | -0.546 | -0.3509 | Yes |
| 26 | KIAA0895L | KIAA0895 like [Source:HGNC Symbol;Acc:HGNC:34408] | 496 | -0.546 | -0.3228 | Yes |
| 27 | SPATS2L | spermatogenesis associated serine rich 2 like [Source:HGNC Symbol;Acc:HGNC:24574] | 499 | -0.561 | -0.2973 | Yes |
| 28 | MUTYH | mutY DNA glycosylase [Source:HGNC Symbol;Acc:HGNC:7527] | 507 | -0.588 | -0.2791 | Yes |
| 29 | NDUFAF4P1 | NADH:ubiquinone oxidoreductase complex assembly factor 4 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:44194] | 511 | -0.619 | -0.2524 | Yes |
| 30 | CEP63 | centrosomal protein 63 [Source:HGNC Symbol;Acc:HGNC:25815] | 512 | -0.620 | -0.2205 | Yes |
| 31 | NMI | N-myc and STAT interactor [Source:HGNC Symbol;Acc:HGNC:7854] | 606 | -2.174 | -0.2684 | Yes |
| 32 | ZBTB18 | zinc finger and BTB domain containing 18 [Source:HGNC Symbol;Acc:HGNC:13030] | 608 | -2.407 | -0.1463 | Yes |
| 33 | PAXIP1-AS2 | PAXIP1 antisense RNA 2 [Source:HGNC Symbol;Acc:HGNC:48958] | 612 | -3.008 | 0.0034 | Yes |
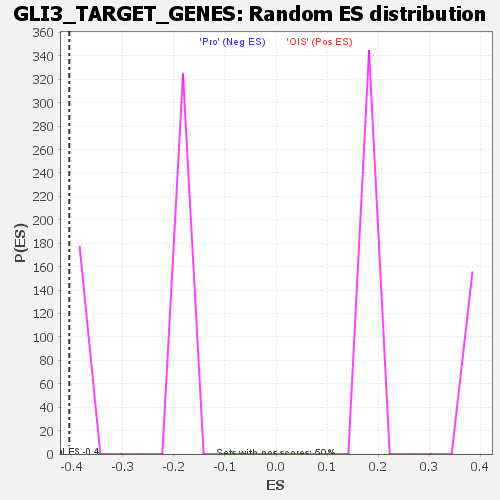
Table: GSEA details [plain text format]