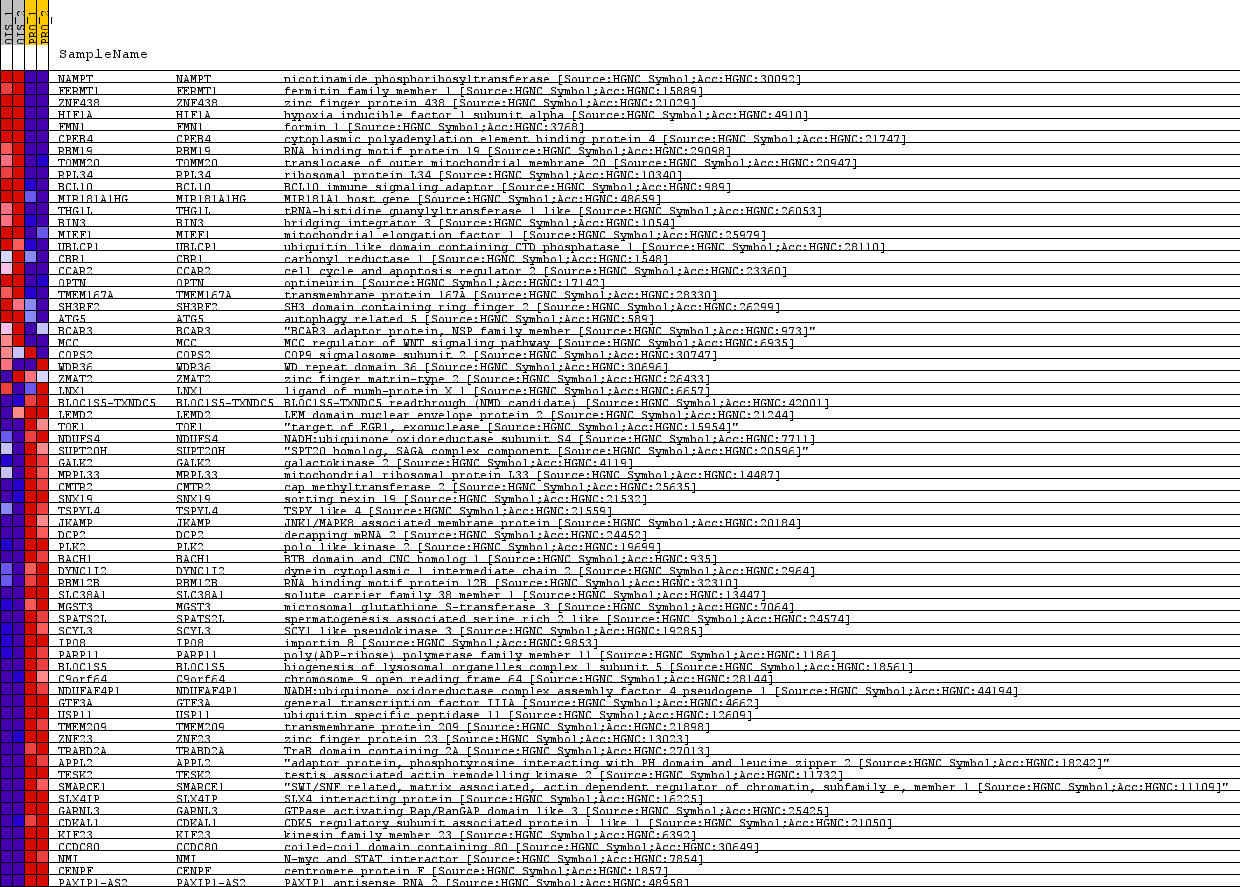
| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | | 1 | NAMPT | nicotinamide phosphoribosyltransferase [Source:HGNC Symbol;Acc:HGNC:30092] | 9 | 4.015 | 0.0645 | No |
| 2 | FERMT1 | fermitin family member 1 [Source:HGNC Symbol;Acc:HGNC:15889] | 38 | 2.479 | 0.0633 | No |
| 3 | ZNF438 | zinc finger protein 438 [Source:HGNC Symbol;Acc:HGNC:21029] | 47 | 2.271 | 0.0945 | No |
| 4 | HIF1A | hypoxia inducible factor 1 subunit alpha [Source:HGNC Symbol;Acc:HGNC:4910] | 83 | 1.244 | 0.0556 | No |
| 5 | FMN1 | formin 1 [Source:HGNC Symbol;Acc:HGNC:3768] | 106 | 0.913 | 0.0338 | No |
| 6 | CPEB4 | cytoplasmic polyadenylation element binding protein 4 [Source:HGNC Symbol;Acc:HGNC:21747] | 118 | 0.788 | 0.0296 | No |
| 7 | RBM19 | RNA binding motif protein 19 [Source:HGNC Symbol;Acc:HGNC:29098] | 123 | 0.757 | 0.0375 | No |
| 8 | TOMM20 | translocase of outer mitochondrial membrane 20 [Source:HGNC Symbol;Acc:HGNC:20947] | 143 | 0.632 | 0.0155 | No |
| 9 | RPL34 | ribosomal protein L34 [Source:HGNC Symbol;Acc:HGNC:10340] | 148 | 0.598 | 0.0203 | No |
| 10 | BCL10 | BCL10 immune signaling adaptor [Source:HGNC Symbol;Acc:HGNC:989] | 156 | 0.568 | 0.0189 | No |
| 11 | MIR181A1HG | MIR181A1 host gene [Source:HGNC Symbol;Acc:HGNC:48659] | 204 | 0.360 | -0.0597 | No |
| 12 | THG1L | tRNA-histidine guanylyltransferase 1 like [Source:HGNC Symbol;Acc:HGNC:26053] | 207 | 0.349 | -0.0563 | No |
| 13 | BIN3 | bridging integrator 3 [Source:HGNC Symbol;Acc:HGNC:1054] | 210 | 0.343 | -0.0531 | No |
| 14 | MIEF1 | mitochondrial elongation factor 1 [Source:HGNC Symbol;Acc:HGNC:25979] | 219 | 0.320 | -0.0613 | No |
| 15 | UBLCP1 | ubiquitin like domain containing CTD phosphatase 1 [Source:HGNC Symbol;Acc:HGNC:28110] | 220 | 0.312 | -0.0550 | No |
| 16 | CBR1 | carbonyl reductase 1 [Source:HGNC Symbol;Acc:HGNC:1548] | 226 | 0.293 | -0.0582 | No |
| 17 | CCAR2 | cell cycle and apoptosis regulator 2 [Source:HGNC Symbol;Acc:HGNC:23360] | 230 | 0.288 | -0.0579 | No |
| 18 | OPTN | optineurin [Source:HGNC Symbol;Acc:HGNC:17142] | 238 | 0.273 | -0.0652 | No |
| 19 | TMEM167A | transmembrane protein 167A [Source:HGNC Symbol;Acc:HGNC:28330] | 257 | 0.213 | -0.0938 | No |
| 20 | SH3RF2 | SH3 domain containing ring finger 2 [Source:HGNC Symbol;Acc:HGNC:26299] | 259 | 0.210 | -0.0914 | No |
| 21 | ATG5 | autophagy related 5 [Source:HGNC Symbol;Acc:HGNC:589] | 273 | 0.178 | -0.1116 | No |
| 22 | BCAR3 | "BCAR3 adaptor protein, NSP family member [Source:HGNC Symbol;Acc:HGNC:973]" | 276 | 0.172 | -0.1117 | No |
| 23 | MCC | MCC regulator of WNT signaling pathway [Source:HGNC Symbol;Acc:HGNC:6935] | 304 | 0.105 | -0.1590 | No |
| 24 | COPS2 | COP9 signalosome subunit 2 [Source:HGNC Symbol;Acc:HGNC:30747] | 349 | 0.001 | -0.2394 | No |
| 25 | WDR36 | WD repeat domain 36 [Source:HGNC Symbol;Acc:HGNC:30696] | 356 | -0.016 | -0.2501 | No |
| 26 | ZMAT2 | zinc finger matrin-type 2 [Source:HGNC Symbol;Acc:HGNC:26433] | 368 | -0.041 | -0.2693 | No |
| 27 | LNX1 | ligand of numb-protein X 1 [Source:HGNC Symbol;Acc:HGNC:6657] | 378 | -0.084 | -0.2841 | No |
| 28 | BLOC1S5-TXNDC5 | BLOC1S5-TXNDC5 readthrough (NMD candidate) [Source:HGNC Symbol;Acc:HGNC:42001] | 382 | -0.114 | -0.2873 | No |
| 29 | LEMD2 | LEM domain nuclear envelope protein 2 [Source:HGNC Symbol;Acc:HGNC:21244] | 385 | -0.128 | -0.2884 | No |
| 30 | TOE1 | "target of EGR1, exonuclease [Source:HGNC Symbol;Acc:HGNC:15954]" | 398 | -0.176 | -0.3068 | No |
| 31 | NDUFS4 | NADH:ubiquinone oxidoreductase subunit S4 [Source:HGNC Symbol;Acc:HGNC:7711] | 400 | -0.197 | -0.3046 | No |
| 32 | SUPT20H | "SPT20 homolog, SAGA complex component [Source:HGNC Symbol;Acc:HGNC:20596]" | 405 | -0.209 | -0.3077 | No |
| 33 | GALK2 | galactokinase 2 [Source:HGNC Symbol;Acc:HGNC:4119] | 410 | -0.238 | -0.3102 | No |
| 34 | MRPL33 | mitochondrial ribosomal protein L33 [Source:HGNC Symbol;Acc:HGNC:14487] | 426 | -0.281 | -0.3320 | No |
| 35 | CMTR2 | cap methyltransferase 2 [Source:HGNC Symbol;Acc:HGNC:25635] | 427 | -0.288 | -0.3262 | No |
| 36 | SNX19 | sorting nexin 19 [Source:HGNC Symbol;Acc:HGNC:21532] | 436 | -0.305 | -0.3347 | No |
| 37 | TSPYL4 | TSPY like 4 [Source:HGNC Symbol;Acc:HGNC:21559] | 451 | -0.366 | -0.3529 | No |
| 38 | JKAMP | JNK1/MAPK8 associated membrane protein [Source:HGNC Symbol;Acc:HGNC:20184] | 453 | -0.373 | -0.3472 | No |
| 39 | DCP2 | decapping mRNA 2 [Source:HGNC Symbol;Acc:HGNC:24452] | 455 | -0.375 | -0.3415 | No |
| 40 | PLK2 | polo like kinase 2 [Source:HGNC Symbol;Acc:HGNC:19699] | 459 | -0.387 | -0.3392 | No |
| 41 | BACH1 | BTB domain and CNC homolog 1 [Source:HGNC Symbol;Acc:HGNC:935] | 476 | -0.450 | -0.3593 | Yes |
| 42 | DYNC1I2 | dynein cytoplasmic 1 intermediate chain 2 [Source:HGNC Symbol;Acc:HGNC:2964] | 479 | -0.465 | -0.3536 | Yes |
| 43 | RBM12B | RNA binding motif protein 12B [Source:HGNC Symbol;Acc:HGNC:32310] | 488 | -0.509 | -0.3580 | Yes |
| 44 | SLC38A1 | solute carrier family 38 member 1 [Source:HGNC Symbol;Acc:HGNC:13447] | 489 | -0.511 | -0.3476 | Yes |
| 45 | MGST3 | microsomal glutathione S-transferase 3 [Source:HGNC Symbol;Acc:HGNC:7064] | 495 | -0.546 | -0.3458 | Yes |
| 46 | SPATS2L | spermatogenesis associated serine rich 2 like [Source:HGNC Symbol;Acc:HGNC:24574] | 499 | -0.561 | -0.3400 | Yes |
| 47 | SCYL3 | SCY1 like pseudokinase 3 [Source:HGNC Symbol;Acc:HGNC:19285] | 501 | -0.574 | -0.3302 | Yes |
| 48 | IPO8 | importin 8 [Source:HGNC Symbol;Acc:HGNC:9853] | 502 | -0.578 | -0.3186 | Yes |
| 49 | PARP11 | poly(ADP-ribose) polymerase family member 11 [Source:HGNC Symbol;Acc:HGNC:1186] | 503 | -0.580 | -0.3068 | Yes |
| 50 | BLOC1S5 | biogenesis of lysosomal organelles complex 1 subunit 5 [Source:HGNC Symbol;Acc:HGNC:18561] | 504 | -0.581 | -0.2951 | Yes |
| 51 | C9orf64 | chromosome 9 open reading frame 64 [Source:HGNC Symbol;Acc:HGNC:28144] | 509 | -0.611 | -0.2901 | Yes |
| 52 | NDUFAF4P1 | NADH:ubiquinone oxidoreductase complex assembly factor 4 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:44194] | 511 | -0.619 | -0.2795 | Yes |
| 53 | GTF3A | general transcription factor IIIA [Source:HGNC Symbol;Acc:HGNC:4662] | 520 | -0.661 | -0.2808 | Yes |
| 54 | USP11 | ubiquitin specific peptidase 11 [Source:HGNC Symbol;Acc:HGNC:12609] | 525 | -0.698 | -0.2740 | Yes |
| 55 | TMEM209 | transmembrane protein 209 [Source:HGNC Symbol;Acc:HGNC:21898] | 528 | -0.717 | -0.2632 | Yes |
| 56 | ZNF23 | zinc finger protein 23 [Source:HGNC Symbol;Acc:HGNC:13023] | 538 | -0.767 | -0.2642 | Yes |
| 57 | TRABD2A | TraB domain containing 2A [Source:HGNC Symbol;Acc:HGNC:27013] | 554 | -0.857 | -0.2743 | Yes |
| 58 | APPL2 | "adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 2 [Source:HGNC Symbol;Acc:HGNC:18242]" | 557 | -0.891 | -0.2600 | Yes |
| 59 | TESK2 | testis associated actin remodelling kinase 2 [Source:HGNC Symbol;Acc:HGNC:11732] | 560 | -0.920 | -0.2451 | Yes |
| 60 | SMARCE1 | "SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 [Source:HGNC Symbol;Acc:HGNC:11109]" | 561 | -0.930 | -0.2264 | Yes |
| 61 | SLX4IP | SLX4 interacting protein [Source:HGNC Symbol;Acc:HGNC:16225] | 562 | -0.943 | -0.2073 | Yes |
| 62 | GARNL3 | GTPase activating Rap/RanGAP domain like 3 [Source:HGNC Symbol;Acc:HGNC:25425] | 576 | -1.082 | -0.2093 | Yes |
| 63 | CDKAL1 | CDK5 regulatory subunit associated protein 1 like 1 [Source:HGNC Symbol;Acc:HGNC:21050] | 578 | -1.112 | -0.1887 | Yes |
| 64 | KIF23 | kinesin family member 23 [Source:HGNC Symbol;Acc:HGNC:6392] | 599 | -1.898 | -0.1870 | Yes |
| 65 | CCDC80 | coiled-coil domain containing 80 [Source:HGNC Symbol;Acc:HGNC:30649] | 604 | -2.118 | -0.1516 | Yes |
| 66 | NMI | N-myc and STAT interactor [Source:HGNC Symbol;Acc:HGNC:7854] | 606 | -2.174 | -0.1095 | Yes |
| 67 | CENPF | centromere protein F [Source:HGNC Symbol;Acc:HGNC:1857] | 611 | -2.967 | -0.0570 | Yes |
| 68 | PAXIP1-AS2 | PAXIP1 antisense RNA 2 [Source:HGNC Symbol;Acc:HGNC:48958] | 612 | -3.008 | 0.0037 | Yes |
Table: GSEA details [plain text format]