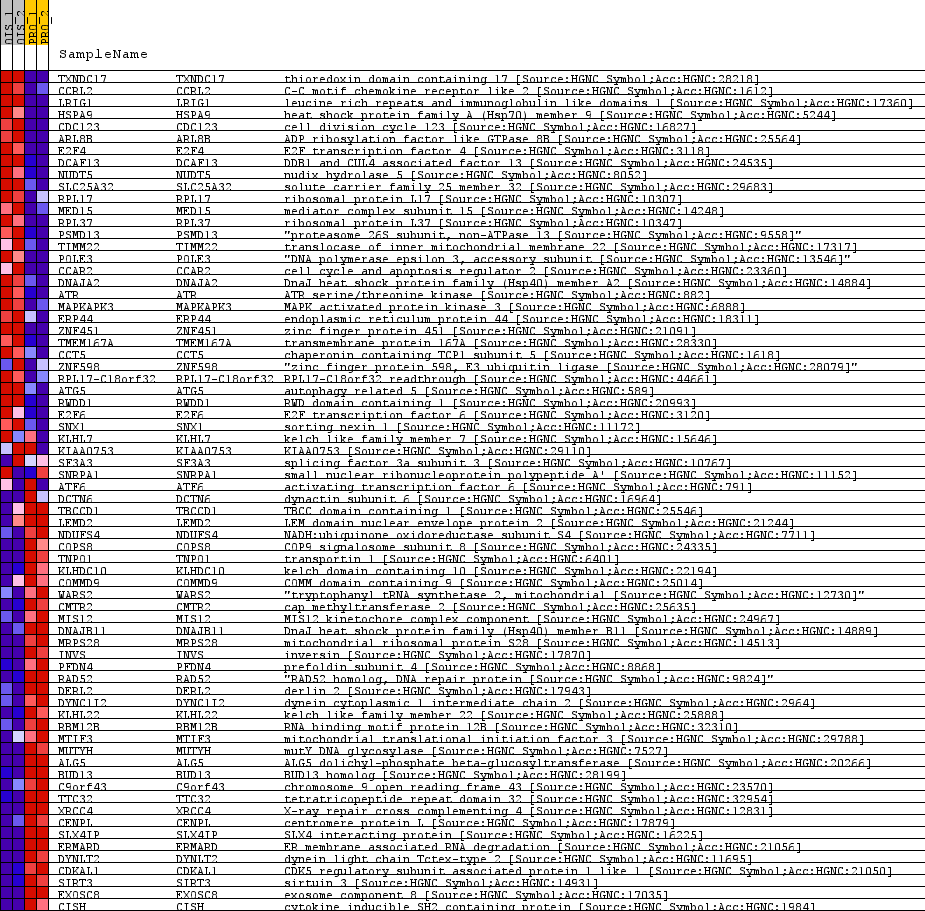
| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | | 1 | TXNDC17 | thioredoxin domain containing 17 [Source:HGNC Symbol;Acc:HGNC:28218] | 74 | 1.441 | -0.0902 | No |
| 2 | CCRL2 | C-C motif chemokine receptor like 2 [Source:HGNC Symbol;Acc:HGNC:1612] | 87 | 1.145 | -0.0760 | No |
| 3 | LRIG1 | leucine rich repeats and immunoglobulin like domains 1 [Source:HGNC Symbol;Acc:HGNC:17360] | 110 | 0.897 | -0.0880 | No |
| 4 | HSPA9 | heat shock protein family A (Hsp70) member 9 [Source:HGNC Symbol;Acc:HGNC:5244] | 127 | 0.744 | -0.0938 | No |
| 5 | CDC123 | cell division cycle 123 [Source:HGNC Symbol;Acc:HGNC:16827] | 146 | 0.607 | -0.1076 | No |
| 6 | ARL8B | ADP ribosylation factor like GTPase 8B [Source:HGNC Symbol;Acc:HGNC:25564] | 173 | 0.460 | -0.1408 | No |
| 7 | E2F4 | E2F transcription factor 4 [Source:HGNC Symbol;Acc:HGNC:3118] | 180 | 0.441 | -0.1378 | No |
| 8 | DCAF13 | DDB1 and CUL4 associated factor 13 [Source:HGNC Symbol;Acc:HGNC:24535] | 183 | 0.434 | -0.1278 | No |
| 9 | NUDT5 | nudix hydrolase 5 [Source:HGNC Symbol;Acc:HGNC:8052] | 187 | 0.415 | -0.1201 | No |
| 10 | SLC25A32 | solute carrier family 25 member 32 [Source:HGNC Symbol;Acc:HGNC:29683] | 193 | 0.401 | -0.1166 | No |
| 11 | RPL17 | ribosomal protein L17 [Source:HGNC Symbol;Acc:HGNC:10307] | 198 | 0.389 | -0.1117 | No |
| 12 | MED15 | mediator complex subunit 15 [Source:HGNC Symbol;Acc:HGNC:14248] | 202 | 0.368 | -0.1055 | No |
| 13 | RPL37 | ribosomal protein L37 [Source:HGNC Symbol;Acc:HGNC:10347] | 212 | 0.340 | -0.1113 | No |
| 14 | PSMD13 | "proteasome 26S subunit, non-ATPase 13 [Source:HGNC Symbol;Acc:HGNC:9558]" | 214 | 0.338 | -0.1024 | No |
| 15 | TIMM22 | translocase of inner mitochondrial membrane 22 [Source:HGNC Symbol;Acc:HGNC:17317] | 217 | 0.324 | -0.0959 | No |
| 16 | POLE3 | "DNA polymerase epsilon 3, accessory subunit [Source:HGNC Symbol;Acc:HGNC:13546]" | 228 | 0.290 | -0.1050 | No |
| 17 | CCAR2 | cell cycle and apoptosis regulator 2 [Source:HGNC Symbol;Acc:HGNC:23360] | 230 | 0.288 | -0.0978 | No |
| 18 | DNAJA2 | DnaJ heat shock protein family (Hsp40) member A2 [Source:HGNC Symbol;Acc:HGNC:14884] | 231 | 0.287 | -0.0887 | No |
| 19 | ATR | ATR serine/threonine kinase [Source:HGNC Symbol;Acc:HGNC:882] | 235 | 0.278 | -0.0854 | No |
| 20 | MAPKAPK3 | MAPK activated protein kinase 3 [Source:HGNC Symbol;Acc:HGNC:6888] | 247 | 0.249 | -0.0977 | No |
| 21 | ERP44 | endoplasmic reticulum protein 44 [Source:HGNC Symbol;Acc:HGNC:18311] | 248 | 0.245 | -0.0899 | No |
| 22 | ZNF451 | zinc finger protein 451 [Source:HGNC Symbol;Acc:HGNC:21091] | 252 | 0.227 | -0.0882 | No |
| 23 | TMEM167A | transmembrane protein 167A [Source:HGNC Symbol;Acc:HGNC:28330] | 257 | 0.213 | -0.0888 | No |
| 24 | CCT5 | chaperonin containing TCP1 subunit 5 [Source:HGNC Symbol;Acc:HGNC:1618] | 260 | 0.210 | -0.0859 | No |
| 25 | ZNF598 | "zinc finger protein 598, E3 ubiquitin ligase [Source:HGNC Symbol;Acc:HGNC:28079]" | 262 | 0.201 | -0.0813 | No |
| 26 | RPL17-C18orf32 | RPL17-C18orf32 readthrough [Source:HGNC Symbol;Acc:HGNC:44661] | 266 | 0.192 | -0.0808 | No |
| 27 | ATG5 | autophagy related 5 [Source:HGNC Symbol;Acc:HGNC:589] | 273 | 0.178 | -0.0862 | No |
| 28 | RWDD1 | RWD domain containing 1 [Source:HGNC Symbol;Acc:HGNC:20993] | 282 | 0.165 | -0.0956 | No |
| 29 | E2F6 | E2F transcription factor 6 [Source:HGNC Symbol;Acc:HGNC:3120] | 296 | 0.120 | -0.1157 | No |
| 30 | SNX1 | sorting nexin 1 [Source:HGNC Symbol;Acc:HGNC:11172] | 297 | 0.120 | -0.1119 | No |
| 31 | KLHL7 | kelch like family member 7 [Source:HGNC Symbol;Acc:HGNC:15646] | 319 | 0.064 | -0.1484 | No |
| 32 | KIAA0753 | KIAA0753 [Source:HGNC Symbol;Acc:HGNC:29110] | 337 | 0.012 | -0.1792 | No |
| 33 | SF3A3 | splicing factor 3a subunit 3 [Source:HGNC Symbol;Acc:HGNC:10767] | 347 | 0.002 | -0.1957 | No |
| 34 | SNRPA1 | small nuclear ribonucleoprotein polypeptide A' [Source:HGNC Symbol;Acc:HGNC:11152] | 350 | 0.000 | -0.1993 | No |
| 35 | ATF6 | activating transcription factor 6 [Source:HGNC Symbol;Acc:HGNC:791] | 355 | -0.015 | -0.2062 | No |
| 36 | DCTN6 | dynactin subunit 6 [Source:HGNC Symbol;Acc:HGNC:16964] | 366 | -0.039 | -0.2233 | No |
| 37 | TBCCD1 | TBCC domain containing 1 [Source:HGNC Symbol;Acc:HGNC:25546] | 383 | -0.115 | -0.2490 | No |
| 38 | LEMD2 | LEM domain nuclear envelope protein 2 [Source:HGNC Symbol;Acc:HGNC:21244] | 385 | -0.128 | -0.2468 | No |
| 39 | NDUFS4 | NADH:ubiquinone oxidoreductase subunit S4 [Source:HGNC Symbol;Acc:HGNC:7711] | 400 | -0.197 | -0.2663 | No |
| 40 | COPS8 | COP9 signalosome subunit 8 [Source:HGNC Symbol;Acc:HGNC:24335] | 415 | -0.252 | -0.2840 | Yes |
| 41 | TNPO1 | transportin 1 [Source:HGNC Symbol;Acc:HGNC:6401] | 417 | -0.257 | -0.2777 | Yes |
| 42 | KLHDC10 | kelch domain containing 10 [Source:HGNC Symbol;Acc:HGNC:22194] | 418 | -0.257 | -0.2695 | Yes |
| 43 | COMMD9 | COMM domain containing 9 [Source:HGNC Symbol;Acc:HGNC:25014] | 419 | -0.258 | -0.2614 | Yes |
| 44 | WARS2 | "tryptophanyl tRNA synthetase 2, mitochondrial [Source:HGNC Symbol;Acc:HGNC:12730]" | 422 | -0.270 | -0.2565 | Yes |
| 45 | CMTR2 | cap methyltransferase 2 [Source:HGNC Symbol;Acc:HGNC:25635] | 427 | -0.288 | -0.2547 | Yes |
| 46 | MIS12 | MIS12 kinetochore complex component [Source:HGNC Symbol;Acc:HGNC:24967] | 445 | -0.348 | -0.2749 | Yes |
| 47 | DNAJB11 | DnaJ heat shock protein family (Hsp40) member B11 [Source:HGNC Symbol;Acc:HGNC:14889] | 446 | -0.348 | -0.2639 | Yes |
| 48 | MRPS28 | mitochondrial ribosomal protein S28 [Source:HGNC Symbol;Acc:HGNC:14513] | 456 | -0.380 | -0.2684 | Yes |
| 49 | INVS | inversin [Source:HGNC Symbol;Acc:HGNC:17870] | 464 | -0.406 | -0.2684 | Yes |
| 50 | PFDN4 | prefoldin subunit 4 [Source:HGNC Symbol;Acc:HGNC:8868] | 467 | -0.415 | -0.2590 | Yes |
| 51 | RAD52 | "RAD52 homolog, DNA repair protein [Source:HGNC Symbol;Acc:HGNC:9824]" | 468 | -0.419 | -0.2457 | Yes |
| 52 | DERL2 | derlin 2 [Source:HGNC Symbol;Acc:HGNC:17943] | 475 | -0.439 | -0.2428 | Yes |
| 53 | DYNC1I2 | dynein cytoplasmic 1 intermediate chain 2 [Source:HGNC Symbol;Acc:HGNC:2964] | 479 | -0.465 | -0.2336 | Yes |
| 54 | KLHL22 | kelch like family member 22 [Source:HGNC Symbol;Acc:HGNC:25888] | 486 | -0.503 | -0.2287 | Yes |
| 55 | RBM12B | RNA binding motif protein 12B [Source:HGNC Symbol;Acc:HGNC:32310] | 488 | -0.509 | -0.2145 | Yes |
| 56 | MTIF3 | mitochondrial translational initiation factor 3 [Source:HGNC Symbol;Acc:HGNC:29788] | 497 | -0.548 | -0.2118 | Yes |
| 57 | MUTYH | mutY DNA glycosylase [Source:HGNC Symbol;Acc:HGNC:7527] | 507 | -0.588 | -0.2097 | Yes |
| 58 | ALG5 | ALG5 dolichyl-phosphate beta-glucosyltransferase [Source:HGNC Symbol;Acc:HGNC:20266] | 510 | -0.613 | -0.1940 | Yes |
| 59 | BUD13 | BUD13 homolog [Source:HGNC Symbol;Acc:HGNC:28199] | 513 | -0.621 | -0.1780 | Yes |
| 60 | C9orf43 | chromosome 9 open reading frame 43 [Source:HGNC Symbol;Acc:HGNC:23570] | 522 | -0.680 | -0.1712 | Yes |
| 61 | TTC32 | tetratricopeptide repeat domain 32 [Source:HGNC Symbol;Acc:HGNC:32954] | 530 | -0.732 | -0.1609 | Yes |
| 62 | XRCC4 | X-ray repair cross complementing 4 [Source:HGNC Symbol;Acc:HGNC:12831] | 533 | -0.745 | -0.1410 | Yes |
| 63 | CENPL | centromere protein L [Source:HGNC Symbol;Acc:HGNC:17879] | 548 | -0.791 | -0.1417 | Yes |
| 64 | SLX4IP | SLX4 interacting protein [Source:HGNC Symbol;Acc:HGNC:16225] | 562 | -0.943 | -0.1357 | Yes |
| 65 | ERMARD | ER membrane associated RNA degradation [Source:HGNC Symbol;Acc:HGNC:21056] | 567 | -1.015 | -0.1109 | Yes |
| 66 | DYNLT2 | dynein light chain Tctex-type 2 [Source:HGNC Symbol;Acc:HGNC:11695] | 573 | -1.044 | -0.0871 | Yes |
| 67 | CDKAL1 | CDK5 regulatory subunit associated protein 1 like 1 [Source:HGNC Symbol;Acc:HGNC:21050] | 578 | -1.112 | -0.0593 | Yes |
| 68 | SIRT3 | sirtuin 3 [Source:HGNC Symbol;Acc:HGNC:14931] | 579 | -1.119 | -0.0239 | Yes |
| 69 | EXOSC8 | exosome component 8 [Source:HGNC Symbol;Acc:HGNC:17035] | 583 | -1.223 | 0.0093 | Yes |
| 70 | CISH | cytokine inducible SH2 containing protein [Source:HGNC Symbol;Acc:HGNC:1984] | 587 | -1.446 | 0.0495 | Yes |
Table: GSEA details [plain text format]