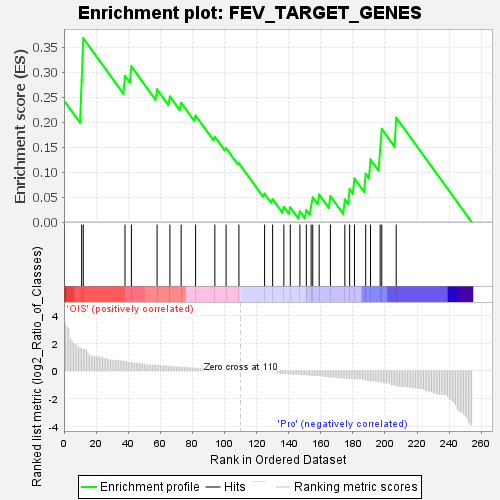
| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | | 1 | HOXB9 | homeobox B9 [Source:HGNC Symbol;Acc:HGNC:5120] | 0 | 4.559 | 0.2431 | Yes |
| 2 | SPRED2 | sprouty related EVH1 domain containing 2 [Source:HGNC Symbol;Acc:HGNC:17722] | 11 | 1.596 | 0.2838 | Yes |
| 3 | ITGB5 | integrin subunit beta 5 [Source:HGNC Symbol;Acc:HGNC:6160] | 12 | 1.580 | 0.3680 | Yes |
| 4 | SNORA33 | "small nucleolar RNA, H/ACA box 33 [Source:HGNC Symbol;Acc:HGNC:32623]" | 38 | 0.669 | 0.2926 | No |
| 5 | SLC26A2 | solute carrier family 26 member 2 [Source:HGNC Symbol;Acc:HGNC:10994] | 42 | 0.604 | 0.3114 | No |
| 6 | CCNJL | cyclin J like [Source:HGNC Symbol;Acc:HGNC:25876] | 58 | 0.388 | 0.2654 | No |
| 7 | DEPTOR | DEP domain containing MTOR interacting protein [Source:HGNC Symbol;Acc:HGNC:22953] | 66 | 0.314 | 0.2510 | No |
| 8 | SHC4 | SHC adaptor protein 4 [Source:HGNC Symbol;Acc:HGNC:16743] | 73 | 0.268 | 0.2386 | No |
| 9 | INTS6 | integrator complex subunit 6 [Source:HGNC Symbol;Acc:HGNC:14879] | 82 | 0.185 | 0.2129 | No |
| 10 | YIPF5 | Yip1 domain family member 5 [Source:HGNC Symbol;Acc:HGNC:24877] | 94 | 0.122 | 0.1706 | No |
| 11 | RFESD | Rieske Fe-S domain containing [Source:HGNC Symbol;Acc:HGNC:29587] | 101 | 0.073 | 0.1478 | No |
| 12 | ZC3H18 | zinc finger CCCH-type containing 18 [Source:HGNC Symbol;Acc:HGNC:25091] | 109 | 0.015 | 0.1174 | No |
| 13 | DUSP10 | dual specificity phosphatase 10 [Source:HGNC Symbol;Acc:HGNC:3065] | 125 | -0.108 | 0.0566 | No |
| 14 | HOXA9 | homeobox A9 [Source:HGNC Symbol;Acc:HGNC:5109] | 130 | -0.128 | 0.0456 | No |
| 15 | SRSF4 | serine and arginine rich splicing factor 4 [Source:HGNC Symbol;Acc:HGNC:10786] | 137 | -0.208 | 0.0301 | No |
| 16 | CCDC112 | coiled-coil domain containing 112 [Source:HGNC Symbol;Acc:HGNC:28599] | 141 | -0.233 | 0.0292 | No |
| 17 | SPOP | speckle type BTB/POZ protein [Source:HGNC Symbol;Acc:HGNC:11254] | 147 | -0.263 | 0.0210 | No |
| 18 | ISCU | iron-sulfur cluster assembly enzyme [Source:HGNC Symbol;Acc:HGNC:29882] | 151 | -0.300 | 0.0237 | No |
| 19 | GFM1 | G elongation factor mitochondrial 1 [Source:HGNC Symbol;Acc:HGNC:13780] | 154 | -0.319 | 0.0318 | No |
| 20 | FAM174B | family with sequence similarity 174 member B [Source:HGNC Symbol;Acc:HGNC:34339] | 155 | -0.327 | 0.0492 | No |
| 21 | KCTD16 | potassium channel tetramerization domain containing 16 [Source:HGNC Symbol;Acc:HGNC:29244] | 159 | -0.353 | 0.0547 | No |
| 22 | PRELID2 | PRELI domain containing 2 [Source:HGNC Symbol;Acc:HGNC:28306] | 166 | -0.449 | 0.0520 | No |
| 23 | TTC23 | tetratricopeptide repeat domain 23 [Source:HGNC Symbol;Acc:HGNC:25730] | 175 | -0.534 | 0.0449 | No |
| 24 | NEK9 | NIMA related kinase 9 [Source:HGNC Symbol;Acc:HGNC:18591] | 178 | -0.554 | 0.0656 | No |
| 25 | OSBPL11 | oxysterol binding protein like 11 [Source:HGNC Symbol;Acc:HGNC:16397] | 181 | -0.569 | 0.0870 | No |
| 26 | LRRC28 | leucine rich repeat containing 28 [Source:HGNC Symbol;Acc:HGNC:28355] | 188 | -0.670 | 0.0961 | No |
| 27 | ARL4A | ADP ribosylation factor like GTPase 4A [Source:HGNC Symbol;Acc:HGNC:695] | 191 | -0.708 | 0.1250 | No |
| 28 | ZCRB1 | zinc finger CCHC-type and RNA binding motif containing 1 [Source:HGNC Symbol;Acc:HGNC:29620] | 197 | -0.772 | 0.1439 | No |
| 29 | EFHC1 | EF-hand domain containing 1 [Source:HGNC Symbol;Acc:HGNC:16406] | 198 | -0.794 | 0.1863 | No |
| 30 | GULP1 | GULP PTB domain containing engulfment adaptor 1 [Source:HGNC Symbol;Acc:HGNC:18649] | 207 | -1.091 | 0.2089 | No |
Table: GSEA details [plain text format]