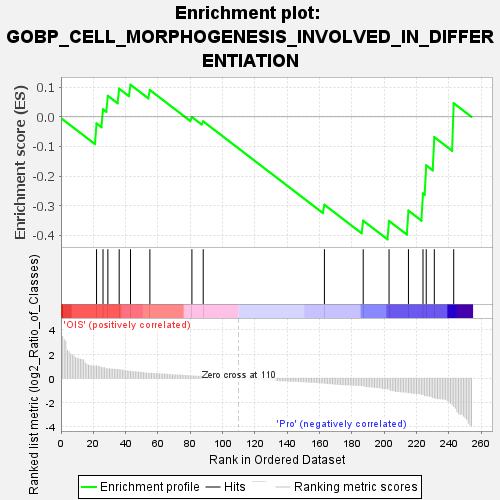
| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | | 1 | EPHB2 | EPH receptor B2 [Source:HGNC Symbol;Acc:HGNC:3393] | 22 | 1.010 | -0.0224 | No |
| 2 | LGR6 | leucine rich repeat containing G protein-coupled receptor 6 [Source:HGNC Symbol;Acc:HGNC:19719] | 26 | 0.877 | 0.0255 | No |
| 3 | NTNG2 | netrin G2 [Source:HGNC Symbol;Acc:HGNC:14288] | 29 | 0.782 | 0.0710 | No |
| 4 | BAIAP2 | BAR/IMD domain containing adaptor protein 2 [Source:HGNC Symbol;Acc:HGNC:947] | 36 | 0.712 | 0.0950 | No |
| 5 | VLDLR | very low density lipoprotein receptor [Source:HGNC Symbol;Acc:HGNC:12698] | 43 | 0.561 | 0.1085 | No |
| 6 | TRPC6 | transient receptor potential cation channel subfamily C member 6 [Source:HGNC Symbol;Acc:HGNC:12338] | 55 | 0.409 | 0.0907 | No |
| 7 | ABL1 | "ABL proto-oncogene 1, non-receptor tyrosine kinase [Source:HGNC Symbol;Acc:HGNC:76]" | 81 | 0.192 | -0.0007 | No |
| 8 | PRKDC | "protein kinase, DNA-activated, catalytic subunit [Source:HGNC Symbol;Acc:HGNC:9413]" | 88 | 0.152 | -0.0153 | No |
| 9 | PARVA | parvin alpha [Source:HGNC Symbol;Acc:HGNC:14652] | 163 | -0.401 | -0.2972 | No |
| 10 | MATN1 | matrilin 1 [Source:HGNC Symbol;Acc:HGNC:6907] | 187 | -0.616 | -0.3510 | No |
| 11 | CDH11 | cadherin 11 [Source:HGNC Symbol;Acc:HGNC:1750] | 203 | -0.898 | -0.3519 | Yes |
| 12 | DOK6 | docking protein 6 [Source:HGNC Symbol;Acc:HGNC:28301] | 215 | -1.172 | -0.3171 | Yes |
| 13 | TRIO | trio Rho guanine nucleotide exchange factor [Source:HGNC Symbol;Acc:HGNC:12303] | 224 | -1.330 | -0.2589 | Yes |
| 14 | BDNF | brain derived neurotrophic factor [Source:HGNC Symbol;Acc:HGNC:1033] | 226 | -1.437 | -0.1641 | Yes |
| 15 | SDC2 | syndecan 2 [Source:HGNC Symbol;Acc:HGNC:10659] | 231 | -1.628 | -0.0686 | Yes |
| 16 | FZD7 | frizzled class receptor 7 [Source:HGNC Symbol;Acc:HGNC:4045] | 243 | -2.331 | 0.0460 | Yes |
Table: GSEA details [plain text format]