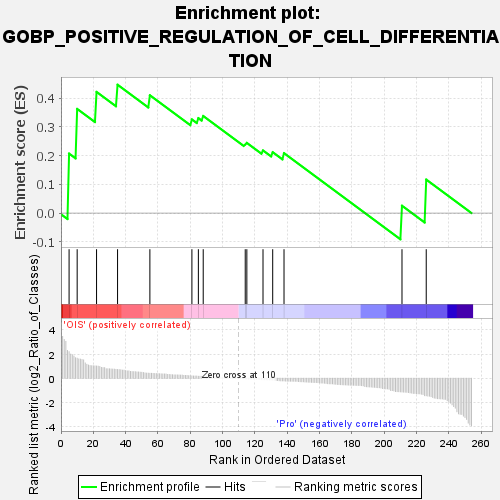
| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | | 1 | FOS | "Fos proto-oncogene, AP-1 transcription factor subunit [Source:HGNC Symbol;Acc:HGNC:3796]" | 5 | 2.193 | 0.2072 | Yes |
| 2 | GREM1 | "gremlin 1, DAN family BMP antagonist [Source:HGNC Symbol;Acc:HGNC:2001]" | 10 | 1.657 | 0.3628 | Yes |
| 3 | EPHB2 | EPH receptor B2 [Source:HGNC Symbol;Acc:HGNC:3393] | 22 | 1.010 | 0.4220 | Yes |
| 4 | PRKCH | protein kinase C eta [Source:HGNC Symbol;Acc:HGNC:9403] | 35 | 0.721 | 0.4470 | Yes |
| 5 | TRPC6 | transient receptor potential cation channel subfamily C member 6 [Source:HGNC Symbol;Acc:HGNC:12338] | 55 | 0.409 | 0.4103 | No |
| 6 | ABL1 | "ABL proto-oncogene 1, non-receptor tyrosine kinase [Source:HGNC Symbol;Acc:HGNC:76]" | 81 | 0.192 | 0.3262 | No |
| 7 | PPARD | peroxisome proliferator activated receptor delta [Source:HGNC Symbol;Acc:HGNC:9235] | 85 | 0.163 | 0.3306 | No |
| 8 | PRKDC | "protein kinase, DNA-activated, catalytic subunit [Source:HGNC Symbol;Acc:HGNC:9413]" | 88 | 0.152 | 0.3381 | No |
| 9 | RGCC | regulator of cell cycle [Source:HGNC Symbol;Acc:HGNC:20369] | 114 | -0.044 | 0.2385 | No |
| 10 | MALT1 | MALT1 paracaspase [Source:HGNC Symbol;Acc:HGNC:6819] | 115 | -0.056 | 0.2444 | No |
| 11 | DUSP10 | dual specificity phosphatase 10 [Source:HGNC Symbol;Acc:HGNC:3065] | 125 | -0.108 | 0.2182 | No |
| 12 | NF2 | neurofibromin 2 [Source:HGNC Symbol;Acc:HGNC:7773] | 131 | -0.142 | 0.2120 | No |
| 13 | NAP1L1 | nucleosome assembly protein 1 like 1 [Source:HGNC Symbol;Acc:HGNC:7637] | 138 | -0.208 | 0.2087 | No |
| 14 | MYOCD | myocardin [Source:HGNC Symbol;Acc:HGNC:16067] | 211 | -1.124 | 0.0256 | No |
| 15 | BDNF | brain derived neurotrophic factor [Source:HGNC Symbol;Acc:HGNC:1033] | 226 | -1.437 | 0.1167 | No |
Table: GSEA details [plain text format]