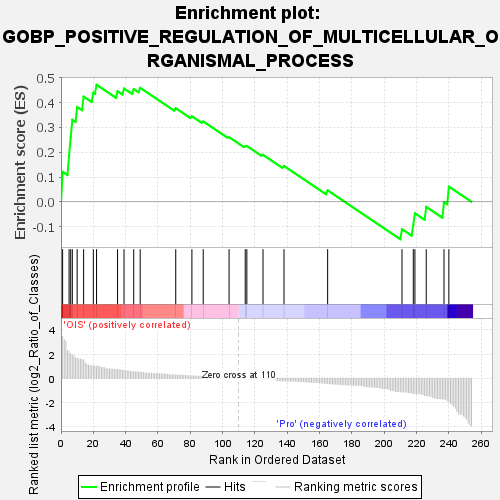
| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | | 1 | NTSR1 | neurotensin receptor 1 [Source:HGNC Symbol;Acc:HGNC:8039] | 1 | 3.452 | 0.1211 | Yes |
| 2 | FOS | "Fos proto-oncogene, AP-1 transcription factor subunit [Source:HGNC Symbol;Acc:HGNC:3796]" | 5 | 2.193 | 0.1876 | Yes |
| 3 | LAPTM5 | lysosomal protein transmembrane 5 [Source:HGNC Symbol;Acc:HGNC:29612] | 6 | 1.996 | 0.2601 | Yes |
| 4 | RGS4 | regulator of G protein signaling 4 [Source:HGNC Symbol;Acc:HGNC:10000] | 7 | 1.947 | 0.3309 | Yes |
| 5 | GREM1 | "gremlin 1, DAN family BMP antagonist [Source:HGNC Symbol;Acc:HGNC:2001]" | 10 | 1.657 | 0.3823 | Yes |
| 6 | POU2AF1 | POU class 2 homeobox associating factor 1 [Source:HGNC Symbol;Acc:HGNC:9211] | 14 | 1.515 | 0.4242 | Yes |
| 7 | FRMD8 | FERM domain containing 8 [Source:HGNC Symbol;Acc:HGNC:25462] | 20 | 1.030 | 0.4397 | Yes |
| 8 | EPHB2 | EPH receptor B2 [Source:HGNC Symbol;Acc:HGNC:3393] | 22 | 1.010 | 0.4721 | Yes |
| 9 | PRKCH | protein kinase C eta [Source:HGNC Symbol;Acc:HGNC:9403] | 35 | 0.721 | 0.4456 | No |
| 10 | POLR3A | RNA polymerase III subunit A [Source:HGNC Symbol;Acc:HGNC:30074] | 39 | 0.669 | 0.4568 | No |
| 11 | PDGFC | platelet derived growth factor C [Source:HGNC Symbol;Acc:HGNC:8801] | 45 | 0.543 | 0.4546 | No |
| 12 | NPAS2 | neuronal PAS domain protein 2 [Source:HGNC Symbol;Acc:HGNC:7895] | 49 | 0.502 | 0.4597 | No |
| 13 | ADORA2A | adenosine A2a receptor [Source:HGNC Symbol;Acc:HGNC:263] | 71 | 0.274 | 0.3775 | No |
| 14 | ABL1 | "ABL proto-oncogene 1, non-receptor tyrosine kinase [Source:HGNC Symbol;Acc:HGNC:76]" | 81 | 0.192 | 0.3450 | No |
| 15 | PRKDC | "protein kinase, DNA-activated, catalytic subunit [Source:HGNC Symbol;Acc:HGNC:9413]" | 88 | 0.152 | 0.3242 | No |
| 16 | SNX5 | sorting nexin 5 [Source:HGNC Symbol;Acc:HGNC:14969] | 104 | 0.066 | 0.2608 | No |
| 17 | RGCC | regulator of cell cycle [Source:HGNC Symbol;Acc:HGNC:20369] | 114 | -0.044 | 0.2229 | No |
| 18 | MALT1 | MALT1 paracaspase [Source:HGNC Symbol;Acc:HGNC:6819] | 115 | -0.056 | 0.2250 | No |
| 19 | DUSP10 | dual specificity phosphatase 10 [Source:HGNC Symbol;Acc:HGNC:3065] | 125 | -0.108 | 0.1895 | No |
| 20 | NAP1L1 | nucleosome assembly protein 1 like 1 [Source:HGNC Symbol;Acc:HGNC:7637] | 138 | -0.208 | 0.1444 | No |
| 21 | FGF1 | fibroblast growth factor 1 [Source:HGNC Symbol;Acc:HGNC:3665] | 165 | -0.438 | 0.0463 | No |
| 22 | MYOCD | myocardin [Source:HGNC Symbol;Acc:HGNC:16067] | 211 | -1.124 | -0.1102 | No |
| 23 | GHRH | growth hormone releasing hormone [Source:HGNC Symbol;Acc:HGNC:4265] | 218 | -1.243 | -0.0914 | No |
| 24 | INHBB | inhibin subunit beta B [Source:HGNC Symbol;Acc:HGNC:6067] | 219 | -1.243 | -0.0462 | No |
| 25 | BDNF | brain derived neurotrophic factor [Source:HGNC Symbol;Acc:HGNC:1033] | 226 | -1.437 | -0.0203 | No |
| 26 | ADRB2 | adrenoceptor beta 2 [Source:HGNC Symbol;Acc:HGNC:286] | 237 | -1.733 | -0.0012 | No |
| 27 | FST | follistatin [Source:HGNC Symbol;Acc:HGNC:3971] | 240 | -1.963 | 0.0614 | No |
Table: GSEA details [plain text format]