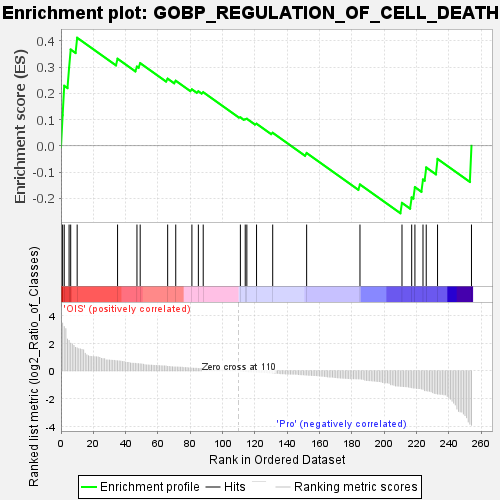
| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | | 1 | NTSR1 | neurotensin receptor 1 [Source:HGNC Symbol;Acc:HGNC:8039] | 1 | 3.452 | 0.1167 | Yes |
| 2 | ECSCR | endothelial cell surface expressed chemotaxis and apoptosis regulator [Source:HGNC Symbol;Acc:HGNC:35454] | 2 | 3.197 | 0.2288 | Yes |
| 3 | FOS | "Fos proto-oncogene, AP-1 transcription factor subunit [Source:HGNC Symbol;Acc:HGNC:3796]" | 5 | 2.193 | 0.2970 | Yes |
| 4 | LAPTM5 | lysosomal protein transmembrane 5 [Source:HGNC Symbol;Acc:HGNC:29612] | 6 | 1.996 | 0.3670 | Yes |
| 5 | GREM1 | "gremlin 1, DAN family BMP antagonist [Source:HGNC Symbol;Acc:HGNC:2001]" | 10 | 1.657 | 0.4119 | Yes |
| 6 | PRKCH | protein kinase C eta [Source:HGNC Symbol;Acc:HGNC:9403] | 35 | 0.721 | 0.3320 | No |
| 7 | SH3RF1 | SH3 domain containing ring finger 1 [Source:HGNC Symbol;Acc:HGNC:17650] | 47 | 0.519 | 0.3019 | No |
| 8 | NPAS2 | neuronal PAS domain protein 2 [Source:HGNC Symbol;Acc:HGNC:7895] | 49 | 0.502 | 0.3151 | No |
| 9 | DEPTOR | DEP domain containing MTOR interacting protein [Source:HGNC Symbol;Acc:HGNC:22953] | 66 | 0.314 | 0.2560 | No |
| 10 | ADORA2A | adenosine A2a receptor [Source:HGNC Symbol;Acc:HGNC:263] | 71 | 0.274 | 0.2480 | No |
| 11 | ABL1 | "ABL proto-oncogene 1, non-receptor tyrosine kinase [Source:HGNC Symbol;Acc:HGNC:76]" | 81 | 0.192 | 0.2153 | No |
| 12 | PPARD | peroxisome proliferator activated receptor delta [Source:HGNC Symbol;Acc:HGNC:9235] | 85 | 0.163 | 0.2079 | No |
| 13 | PRKDC | "protein kinase, DNA-activated, catalytic subunit [Source:HGNC Symbol;Acc:HGNC:9413]" | 88 | 0.152 | 0.2044 | No |
| 14 | BAG4 | BAG cochaperone 4 [Source:HGNC Symbol;Acc:HGNC:940] | 111 | -0.009 | 0.1082 | No |
| 15 | RGCC | regulator of cell cycle [Source:HGNC Symbol;Acc:HGNC:20369] | 114 | -0.044 | 0.1010 | No |
| 16 | MALT1 | MALT1 paracaspase [Source:HGNC Symbol;Acc:HGNC:6819] | 115 | -0.056 | 0.1030 | No |
| 17 | PPID | peptidylprolyl isomerase D [Source:HGNC Symbol;Acc:HGNC:9257] | 121 | -0.086 | 0.0841 | No |
| 18 | NF2 | neurofibromin 2 [Source:HGNC Symbol;Acc:HGNC:7773] | 131 | -0.142 | 0.0496 | No |
| 19 | GRK5 | G protein-coupled receptor kinase 5 [Source:HGNC Symbol;Acc:HGNC:4544] | 152 | -0.301 | -0.0276 | No |
| 20 | KLF11 | Kruppel like factor 11 [Source:HGNC Symbol;Acc:HGNC:11811] | 185 | -0.583 | -0.1475 | No |
| 21 | MYOCD | myocardin [Source:HGNC Symbol;Acc:HGNC:16067] | 211 | -1.124 | -0.2177 | No |
| 22 | MECOM | MDS1 and EVI1 complex locus [Source:HGNC Symbol;Acc:HGNC:3498] | 217 | -1.217 | -0.1970 | No |
| 23 | INHBB | inhibin subunit beta B [Source:HGNC Symbol;Acc:HGNC:6067] | 219 | -1.243 | -0.1578 | No |
| 24 | TRIO | trio Rho guanine nucleotide exchange factor [Source:HGNC Symbol;Acc:HGNC:12303] | 224 | -1.330 | -0.1287 | No |
| 25 | BDNF | brain derived neurotrophic factor [Source:HGNC Symbol;Acc:HGNC:1033] | 226 | -1.437 | -0.0826 | No |
| 26 | CTSC | cathepsin C [Source:HGNC Symbol;Acc:HGNC:2528] | 233 | -1.677 | -0.0501 | No |
| 27 | RASSF2 | Ras association domain family member 2 [Source:HGNC Symbol;Acc:HGNC:9883] | 254 | -3.930 | -0.0000 | No |
Table: GSEA details [plain text format]