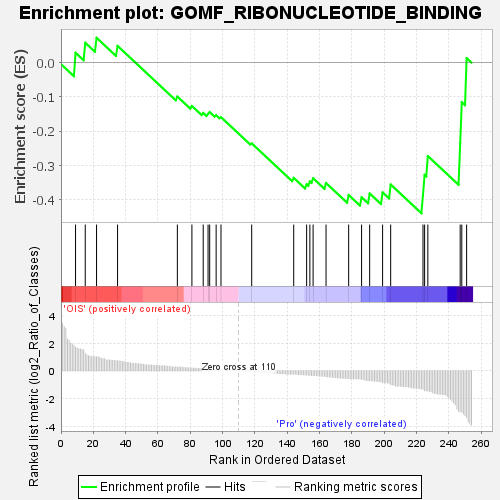
| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | | 1 | RAB31 | "RAB31, member RAS oncogene family [Source:HGNC Symbol;Acc:HGNC:9771]" | 9 | 1.692 | 0.0288 | No |
| 2 | SCG5 | secretogranin V [Source:HGNC Symbol;Acc:HGNC:10816] | 15 | 1.265 | 0.0580 | No |
| 3 | EPHB2 | EPH receptor B2 [Source:HGNC Symbol;Acc:HGNC:3393] | 22 | 1.010 | 0.0725 | No |
| 4 | PRKCH | protein kinase C eta [Source:HGNC Symbol;Acc:HGNC:9403] | 35 | 0.721 | 0.0488 | No |
| 5 | UBA5 | ubiquitin like modifier activating enzyme 5 [Source:HGNC Symbol;Acc:HGNC:23230] | 72 | 0.272 | -0.0988 | No |
| 6 | ABL1 | "ABL proto-oncogene 1, non-receptor tyrosine kinase [Source:HGNC Symbol;Acc:HGNC:76]" | 81 | 0.192 | -0.1262 | No |
| 7 | PRKDC | "protein kinase, DNA-activated, catalytic subunit [Source:HGNC Symbol;Acc:HGNC:9413]" | 88 | 0.152 | -0.1465 | No |
| 8 | BBS10 | Bardet-Biedl syndrome 10 [Source:HGNC Symbol;Acc:HGNC:26291] | 91 | 0.146 | -0.1494 | No |
| 9 | SCYL1 | SCY1 like pseudokinase 1 [Source:HGNC Symbol;Acc:HGNC:14372] | 92 | 0.131 | -0.1441 | No |
| 10 | DGKH | diacylglycerol kinase eta [Source:HGNC Symbol;Acc:HGNC:2854] | 96 | 0.101 | -0.1532 | No |
| 11 | TUT1 | "terminal uridylyl transferase 1, U6 snRNA-specific [Source:HGNC Symbol;Acc:HGNC:26184]" | 99 | 0.077 | -0.1589 | No |
| 12 | DYRK2 | dual specificity tyrosine phosphorylation regulated kinase 2 [Source:HGNC Symbol;Acc:HGNC:3093] | 118 | -0.074 | -0.2352 | No |
| 13 | IFI44L | interferon induced protein 44 like [Source:HGNC Symbol;Acc:HGNC:17817] | 144 | -0.238 | -0.3357 | No |
| 14 | GRK5 | G protein-coupled receptor kinase 5 [Source:HGNC Symbol;Acc:HGNC:4544] | 152 | -0.301 | -0.3544 | No |
| 15 | GFM1 | G elongation factor mitochondrial 1 [Source:HGNC Symbol;Acc:HGNC:13780] | 154 | -0.319 | -0.3459 | No |
| 16 | TTLL11 | tubulin tyrosine ligase like 11 [Source:HGNC Symbol;Acc:HGNC:18113] | 156 | -0.335 | -0.3368 | No |
| 17 | CDK6 | cyclin dependent kinase 6 [Source:HGNC Symbol;Acc:HGNC:1777] | 164 | -0.414 | -0.3508 | No |
| 18 | NEK9 | NIMA related kinase 9 [Source:HGNC Symbol;Acc:HGNC:18591] | 178 | -0.554 | -0.3857 | No |
| 19 | MCM8 | minichromosome maintenance 8 homologous recombination repair factor [Source:HGNC Symbol;Acc:HGNC:16147] | 186 | -0.600 | -0.3922 | Yes |
| 20 | ARL4A | ADP ribosylation factor like GTPase 4A [Source:HGNC Symbol;Acc:HGNC:695] | 191 | -0.708 | -0.3812 | Yes |
| 21 | CHD2 | chromodomain helicase DNA binding protein 2 [Source:HGNC Symbol;Acc:HGNC:1917] | 199 | -0.840 | -0.3780 | Yes |
| 22 | ME1 | malic enzyme 1 [Source:HGNC Symbol;Acc:HGNC:6983] | 204 | -1.002 | -0.3551 | Yes |
| 23 | TRIO | trio Rho guanine nucleotide exchange factor [Source:HGNC Symbol;Acc:HGNC:12303] | 224 | -1.330 | -0.3849 | Yes |
| 24 | HSP90B2P | "heat shock protein 90 beta family member 2, pseudogene [Source:HGNC Symbol;Acc:HGNC:12099]" | 225 | -1.431 | -0.3270 | Yes |
| 25 | MCM4 | minichromosome maintenance complex component 4 [Source:HGNC Symbol;Acc:HGNC:6947] | 227 | -1.463 | -0.2722 | Yes |
| 26 | CDK15 | cyclin dependent kinase 15 [Source:HGNC Symbol;Acc:HGNC:14434] | 247 | -2.968 | -0.2358 | Yes |
| 27 | KIF18A | kinesin family member 18A [Source:HGNC Symbol;Acc:HGNC:29441] | 248 | -2.979 | -0.1152 | Yes |
| 28 | ROR1 | receptor tyrosine kinase like orphan receptor 1 [Source:HGNC Symbol;Acc:HGNC:10256] | 251 | -3.389 | 0.0132 | Yes |
Table: GSEA details [plain text format]