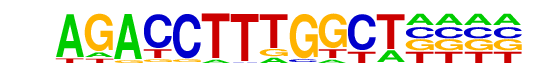
| p-value: | 1e-17 |
| log p-value: | -4.131e+01 |
| Information Content per bp: | 1.743 |
| Number of Target Sequences with motif | 18.0 |
| Percentage of Target Sequences with motif | 9.14% |
| Number of Background Sequences with motif | 1.3 |
| Percentage of Background Sequences with motif | 0.55% |
| Average Position of motif in Targets | 157.0 +/- 92.0bp |
| Average Position of motif in Background | 66.1 +/- 10.4bp |
| Strand Bias (log2 ratio + to - strand density) | 0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.06 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
Erra(NR)/HepG2-Erra-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 1 |
| Score: | 0.69
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -AGACCTTTGGCT
CTGACCTTTG--- |
|


|
|
MA0512.1_Rxra/Jaspar
| Match Rank: | 2 |
| Score: | 0.68
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --AGACCTTTGGCT
NCTGACCTTTG--- |
|


|
|
PB0053.1_Rara_1/Jaspar
| Match Rank: | 3 |
| Score: | 0.67
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----AGACCTTTGGCT
NNNGTGACCTTTGNNN |
|


|
|
Nur77(NR)/K562-NR4A1-ChIP-Seq(GSE31363)/Homer
| Match Rank: | 4 |
| Score: | 0.66
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AGACCTTTGGCT
TGACCTTTNCNT |
|


|
|
PB0049.1_Nr2f2_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.66
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----AGACCTTTGGCT
NNNNTGACCTTTNNNN |
|


|
|
MA0514.1_Sox3/Jaspar
| Match Rank: | 6 |
| Score: | 0.66
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | AGACCTTTGGCT-
---CCTTTGTTTT |
|


|
|
PPARE(NR/DR1)/3T3L1-Pparg-ChIP-Seq(GSE13511)/Homer
| Match Rank: | 7 |
| Score: | 0.65
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AGACCTTTGGCT--
TGACCTTTGCCCCA |
|


|
|
MA0592.1_ESRRA/Jaspar
| Match Rank: | 8 |
| Score: | 0.65
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --AGACCTTTGGCT
NGTGACCTTGG--- |
|


|
|
MA0141.2_Esrrb/Jaspar
| Match Rank: | 9 |
| Score: | 0.64
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | AGACCTTTGGCT
TGACCTTGANNN |
|


|
|
PB0166.1_Sox12_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.64
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | AGACCTTTGGCT----
ANTCCTTTGTCTNNNN |
|
|
|





















