| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | Max(HLH)/K562-Max-ChIP-Seq(GSE31477)/Homer | 1e-4 | -1.102e+01 | 0.0040 | 24.0 | 12.18% | 11.9 | 4.97% | motif file (matrix) |
pdf |
| 2 |  | n-Myc(HLH)/mES-nMyc-ChIP-Seq(GSE11431)/Homer | 1e-4 | -1.031e+01 | 0.0041 | 22.0 | 11.17% | 10.8 | 4.54% | motif file (matrix) |
pdf |
| 3 |  | CEBP:AP1(bZIP)/ThioMac-CEBPb-ChIP-Seq(GSE21512)/Homer | 1e-3 | -8.801e+00 | 0.0124 | 28.0 | 14.21% | 16.6 | 6.94% | motif file (matrix) |
pdf |
| 4 |  | Chop(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer | 1e-3 | -8.375e+00 | 0.0142 | 10.0 | 5.08% | 3.1 | 1.31% | motif file (matrix) |
pdf |
| 5 |  | IRF2(IRF)/Erythroblas-IRF2-ChIP-Seq(GSE36985)/Homer | 1e-3 | -8.119e+00 | 0.0146 | 8.0 | 4.06% | 2.5 | 1.05% | motif file (matrix) |
pdf |
| 6 |  | Oct4(POU/Homeobox)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-3 | -7.821e+00 | 0.0165 | 17.0 | 8.63% | 8.9 | 3.73% | motif file (matrix) |
pdf |
| 7 |  | Atf4(bZIP)/MEF-Atf4-ChIP-Seq(GSE35681)/Homer | 1e-2 | -6.861e+00 | 0.0368 | 12.0 | 6.09% | 5.5 | 2.30% | motif file (matrix) |
pdf |
| 8 |  | NF1(CTF)/LNCAP-NF1-ChIP-Seq(Unpublished)/Homer | 1e-2 | -6.537e+00 | 0.0396 | 17.0 | 8.63% | 9.6 | 4.01% | motif file (matrix) |
pdf |
| 9 |  | Pax8(Paired/Homeobox)/Thyroid-Pax8-ChIP-Seq(GSE26938)/Homer | 1e-2 | -6.537e+00 | 0.0396 | 17.0 | 8.63% | 9.5 | 3.99% | motif file (matrix) |
pdf |
| 10 | 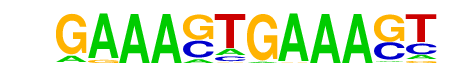 | IRF1(IRF)/PBMC-IRF1-ChIP-Seq(GSE43036)/Homer | 1e-2 | -6.490e+00 | 0.0396 | 7.0 | 3.55% | 2.9 | 1.21% | motif file (matrix) |
pdf |
| 11 |  | PAX3:FKHR-fusion(Paired/Homeobox)/Rh4-PAX3:FKHR-ChIP-Seq(GSE19063)/Homer | 1e-2 | -6.490e+00 | 0.0396 | 7.0 | 3.55% | 2.6 | 1.08% | motif file (matrix) |
pdf |
| 12 |  | HRE(HSF)/Striatum-HSF1-ChIP-Seq(GSE38000)/Homer | 1e-2 | -6.451e+00 | 0.0396 | 5.0 | 2.54% | 1.0 | 0.42% | motif file (matrix) |
pdf |
| 13 |  | Pax7-longest(Paired/Homeobox)/Myoblast-Pax7-ChIP-Seq(GSE25064)/Homer | 1e-2 | -6.451e+00 | 0.0396 | 5.0 | 2.54% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 14 |  | Pbx3(Homeobox)/GM12878-PBX3-ChIP-Seq(GSE32465)/Homer | 1e-2 | -6.451e+00 | 0.0396 | 5.0 | 2.54% | 1.8 | 0.75% | motif file (matrix) |
pdf |
| 15 |  | bHLHE40(HLH)/HepG2-BHLHE40-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.203e+00 | 0.0396 | 10.0 | 5.08% | 4.0 | 1.68% | motif file (matrix) |
pdf |
| 16 |  | Smad3(MAD)/NPC-Smad3-ChIP-Seq(GSE36673)/Homer | 1e-2 | -6.152e+00 | 0.0396 | 86.0 | 43.65% | 80.8 | 33.87% | motif file (matrix) |
pdf |
| 17 |  | c-Myc(HLH)/LNCAP-cMyc-ChIP-Seq(unpublished)/Homer | 1e-2 | -6.090e+00 | 0.0396 | 14.0 | 7.11% | 8.0 | 3.35% | motif file (matrix) |
pdf |
| 18 |  | c-Myc/mES-cMyc-ChIP-Seq(GSE11431)/Homer | 1e-2 | -5.628e+00 | 0.0491 | 16.0 | 8.12% | 9.6 | 4.02% | motif file (matrix) |
pdf |
| 19 |  | CHR/Cell-Cycle-Exp/Homer | 1e-2 | -5.398e+00 | 0.0586 | 29.0 | 14.72% | 21.1 | 8.84% | motif file (matrix) |
pdf |
| 20 |  | NFY(CCAAT)/Promoter/Homer | 1e-2 | -5.127e+00 | 0.0730 | 20.0 | 10.15% | 13.9 | 5.84% | motif file (matrix) |
pdf |
| 21 | 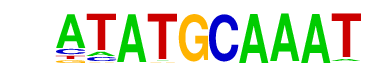 | Oct2(POU/Homeobox)/Bcell-Oct2-ChIP-Seq(GSE21512)/Homer | 1e-2 | -4.673e+00 | 0.1094 | 10.0 | 5.08% | 5.1 | 2.12% | motif file (matrix) |
pdf |









































