| p-value: | 1e-3 |
| log p-value: | -7.724e+00 |
| Information Content per bp: | 1.776 |
| Number of Target Sequences with motif | 422.0 |
| Percentage of Target Sequences with motif | 7.80% |
| Number of Background Sequences with motif | 1180.4 |
| Percentage of Background Sequences with motif | 6.64% |
| Average Position of motif in Targets | 353.7 +/- 342.9bp |
| Average Position of motif in Background | 434.7 +/- 393.8bp |
| Strand Bias (log2 ratio + to - strand density) | -0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.03 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PB0192.1_Tcfap2e_2/Jaspar
| Match Rank: | 1 |
| Score: | 0.82
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CTAAAAAAAA--
TACTGGAAAAAAAA |
|


|
|
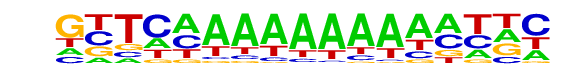
PB0116.1_Elf3_2/Jaspar
| Match Rank: | 2 |
| Score: | 0.78
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---CTAAAAAAAA----
GTTCAAAAAAAAAATTC |
|

|
|
PB0182.1_Srf_2/Jaspar
| Match Rank: | 3 |
| Score: | 0.75
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CTAAAAAAAA------
GTTAAAAAAAAAAATTA |
|


|
|
HOXD13(Homeobox)/Chicken-Hoxd13-ChIP-Seq(GSE38910)/Homer
| Match Rank: | 4 |
| Score: | 0.73
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CTAAAAAAAA
NCYAATAAAA- |
|


|
|
PH0057.1_Hoxb13/Jaspar
| Match Rank: | 5 |
| Score: | 0.73
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---CTAAAAAAAA---
AACCCAATAAAATTCG |
|


|
|
PH0078.1_Hoxd13/Jaspar
| Match Rank: | 6 |
| Score: | 0.72
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---CTAAAAAAAA---
CTACCAATAAAATTCT |
|


|
|
PB0186.1_Tcf3_2/Jaspar
| Match Rank: | 7 |
| Score: | 0.71
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---CTAAAAAAAA--
AGCCGAAAAAAAAAT |
|


|
|
PH0068.1_Hoxc13/Jaspar
| Match Rank: | 8 |
| Score: | 0.70
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----CTAAAAAAAA--
AAAGCTCGTAAAATTT |
|


|
|
PB0148.1_Mtf1_2/Jaspar
| Match Rank: | 9 |
| Score: | 0.70
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CTAAAAAAAA--
AAATAAGAAAAAAC |
|


|
|
PH0048.1_Hoxa13/Jaspar
| Match Rank: | 10 |
| Score: | 0.68
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----CTAAAAAAAA--
AAACCTCGTAAAATTT |
|


|
|





















