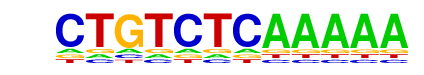











| p-value: | 1e-3 |
| log p-value: | -7.724e+00 |
| Information Content per bp: | 1.776 |
| Number of Target Sequences with motif | 422.0 |
| Percentage of Target Sequences with motif | 7.80% |
| Number of Background Sequences with motif | 1180.4 |
| Percentage of Background Sequences with motif | 6.64% |
| Average Position of motif in Targets | 353.7 +/- 342.9bp |
| Average Position of motif in Background | 434.7 +/- 393.8bp |
| Strand Bias (log2 ratio + to - strand density) | -0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.03 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.663 |  | 1e-2 | -6.820728 | 3.10% | 2.43% | motif file (matrix) |
| 2 | 0.614 |  | 1e-1 | -3.412587 | 1.39% | 1.11% | motif file (matrix) |
| 3 | 0.828 |  | 1e-1 | -2.567442 | 13.77% | 13.10% | motif file (matrix) |
| 4 | 0.701 |  | 1e0 | -1.625344 | 7.17% | 6.87% | motif file (matrix) |
| 5 | 0.712 |  | 1e0 | -0.933816 | 2.40% | 2.34% | motif file (matrix) |
| 6 | 0.608 |  | 1e0 | -0.806349 | 14.56% | 14.49% | motif file (matrix) |
| 7 | 0.692 |  | 1e0 | -0.685975 | 10.53% | 10.53% | motif file (matrix) |
| 8 | 0.763 |  | 1e0 | -0.228084 | 14.30% | 14.70% | motif file (matrix) |
| 9 | 0.848 |  | 1e0 | -0.158138 | 21.64% | 22.23% | motif file (matrix) |
| 10 | 0.808 |  | 1e0 | -0.058559 | 15.34% | 16.12% | motif file (matrix) |