| p-value: | 1e-1 |
| log p-value: | -3.331e+00 |
| Information Content per bp: | 1.883 |
| Number of Target Sequences with motif | 469.0 |
| Percentage of Target Sequences with motif | 8.67% |
| Number of Background Sequences with motif | 1420.4 |
| Percentage of Background Sequences with motif | 7.99% |
| Average Position of motif in Targets | 401.7 +/- 397.2bp |
| Average Position of motif in Background | 453.0 +/- 437.4bp |
| Strand Bias (log2 ratio + to - strand density) | 0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.13 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
MA0019.1_Ddit3::Cebpa/Jaspar
| Match Rank: | 1 |
| Score: | 0.77
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---YGCAATCT-
AGATGCAATCCC |
|


|
|
MF0006.1_bZIP_cEBP-like_subclass/Jaspar
| Match Rank: | 2 |
| Score: | 0.72
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---YGCAATCT
TTATGCAAT-- |
|


|
|
MA0466.1_CEBPB/Jaspar
| Match Rank: | 3 |
| Score: | 0.71
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----YGCAATCT
ATTGTGCAATA- |
|


|
|
MA0102.3_CEBPA/Jaspar
| Match Rank: | 4 |
| Score: | 0.68
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----YGCAATCT
NATTGTGCAAT-- |
|


|
|
MA0038.1_Gfi1/Jaspar
| Match Rank: | 5 |
| Score: | 0.66
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | YGCAATCT---
-CAAATCACTG |
|


|
|
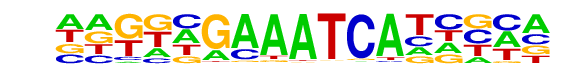
PH0037.1_Hdx/Jaspar
| Match Rank: | 6 |
| Score: | 0.65
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----YGCAATCT-----
AAGGCGAAATCATCGCA |
|

|
|
CEBP(bZIP)/CEBPb-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 7 |
| Score: | 0.63
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----YGCAATCT
GTTGCGCAAT-- |
|


|
|
CEBP:AP1(bZIP)/ThioMac-CEBPb-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 8 |
| Score: | 0.62
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----YGCAATCT
NATGTTGCAA--- |
|


|
|
PB0146.1_Mafk_2/Jaspar
| Match Rank: | 9 |
| Score: | 0.61
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---YGCAATCT----
CCTTGCAATTTTTNN |
|


|
|
Atf4(bZIP)/MEF-Atf4-ChIP-Seq(GSE35681)/Homer
| Match Rank: | 10 |
| Score: | 0.61
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----YGCAATCT
MTGATGCAAT-- |
|


|
|





















