| p-value: | 1e0 |
| log p-value: | -2.680e-01 |
| Information Content per bp: | 1.882 |
| Number of Target Sequences with motif | 674.0 |
| Percentage of Target Sequences with motif | 12.46% |
| Number of Background Sequences with motif | 2271.4 |
| Percentage of Background Sequences with motif | 12.78% |
| Average Position of motif in Targets | 394.0 +/- 365.1bp |
| Average Position of motif in Background | 472.8 +/- 414.7bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.14 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer
| Match Rank: | 1 |
| Score: | 0.79
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -AAGGAATS-
NCTGGAATGC |
|


|
|
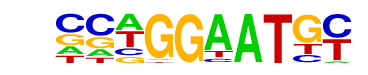
TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer
| Match Rank: | 2 |
| Score: | 0.76
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -AAGGAATS-
CCWGGAATGY |
|

|
|
MA0090.1_TEAD1/Jaspar
| Match Rank: | 3 |
| Score: | 0.72
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --AAGGAATS--
CNGAGGAATGTG |
|


|
|
MA0136.1_ELF5/Jaspar
| Match Rank: | 4 |
| Score: | 0.69
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | AAGGAATS-
AAGGAAGTA |
|


|
|
PB0203.1_Zfp691_2/Jaspar
| Match Rank: | 5 |
| Score: | 0.68
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----AAGGAATS-----
NTNNNAGGAGTCTCNTN |
|


|
|
MA0081.1_SPIB/Jaspar
| Match Rank: | 6 |
| Score: | 0.66
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -AAGGAATS
AGAGGAA-- |
|


|
|
PB0028.1_Hbp1_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.63
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------AAGGAATS--
ACTATGAATGAATGAT |
|


|
|
MA0109.1_Hltf/Jaspar
| Match Rank: | 8 |
| Score: | 0.61
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----AAGGAATS
NNATAAGGNN-- |
|


|
|
PB0166.1_Sox12_2/Jaspar
| Match Rank: | 9 |
| Score: | 0.61
| | Offset: | -9
| | Orientation: | forward strand |
| Alignment: | ---------AAGGAATS
AAACAGACAAAGGAAT- |
|


|
|
MA0598.1_EHF/Jaspar
| Match Rank: | 10 |
| Score: | 0.60
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | AAGGAATS
CAGGAAGG |
|


|
|





















