| p-value: | 1e-15 |
| log p-value: | -3.484e+01 |
| Information Content per bp: | 1.835 |
| Number of Target Sequences with motif | 13.0 |
| Percentage of Target Sequences with motif | 1.30% |
| Number of Background Sequences with motif | 1.1 |
| Percentage of Background Sequences with motif | 0.04% |
| Average Position of motif in Targets | 364.0 +/- 283.0bp |
| Average Position of motif in Background | 720.0 +/- 0.0bp |
| Strand Bias (log2 ratio + to - strand density) | 0.7 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
MA0091.1_TAL1::TCF3/Jaspar
| Match Rank: | 1 |
| Score: | 0.69
| | Offset: | -4
| | Orientation: | forward strand |
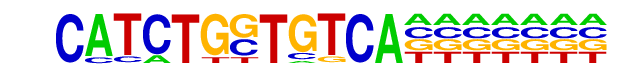
| Alignment: | ----CATCTGSTGTCA
CGACCATCTGTT---- |
|


|
|
NeuroD1(bHLH)/Islet-NeuroD1-ChIP-Seq(GSE30298)/Homer
| Match Rank: | 2 |
| Score: | 0.69
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CATCTGSTGTCA
GCCATCTGTT---- |
|


|
|
Atoh1(bHLH)/Cerebellum-Atoh1-ChIP-Seq(GSE22111)/Homer
| Match Rank: | 3 |
| Score: | 0.66
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CATCTGSTGTCA
GCCAGCTGBTNB-- |
|


|
|
Olig2(bHLH)/Neuron-Olig2-ChIP-Seq(GSE30882)/Homer
| Match Rank: | 4 |
| Score: | 0.66
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CATCTGSTGTCA
RCCATMTGTT---- |
|


|
|
PB0193.1_Tcfe2a_2/Jaspar
| Match Rank: | 5 |
| Score: | 0.65
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------CATCTGSTGTCA
CCNNACCATCTGGCCTN- |
|


|
|
PH0164.1_Six4/Jaspar
| Match Rank: | 6 |
| Score: | 0.64
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CATCTGSTGTCA-----
TNNNNGGTGTCATNTNT |
|


|
|
SCL(HLH)/HPC7-Scl-ChIP-Seq(GSE13511)/Homer
| Match Rank: | 7 |
| Score: | 0.63
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CATCTGSTGTCA
CAGCTGNT---- |
|


|
|
MA0498.1_Meis1/Jaspar
| Match Rank: | 8 |
| Score: | 0.62
| | Offset: | 4
| | Orientation: | forward strand |
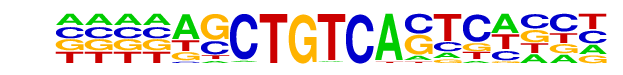
| Alignment: | CATCTGSTGTCA-------
----AGCTGTCACTCACCT |
|
|
|
Tcf12(HLH)/GM12878-Tcf12-ChIP-Seq(GSE32465)/Homer
| Match Rank: | 9 |
| Score: | 0.62
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CATCTGSTGTCA
NCAGCTGCTG--- |
|


|
|
CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer
| Match Rank: | 10 |
| Score: | 0.62
| | Offset: | -7
| | Orientation: | forward strand |
| Alignment: | -------CATCTGSTGTCA-
ANAGTGCCACCTGGTGGCCA |
|


|
|





















