| p-value: | 1e-13 |
| log p-value: | -3.078e+01 |
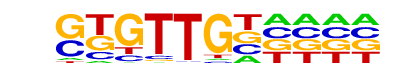
| Information Content per bp: | 1.840 |
| Number of Target Sequences with motif | 28.0 |
| Percentage of Target Sequences with motif | 2.80% |
| Number of Background Sequences with motif | 11.5 |
| Percentage of Background Sequences with motif | 0.46% |
| Average Position of motif in Targets | 305.7 +/- 253.8bp |
| Average Position of motif in Background | 252.1 +/- 217.4bp |
| Strand Bias (log2 ratio + to - strand density) | -0.3 |
| Multiplicity (# of sites on avg that occur together) | 1.04 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
CEBP:AP1(bZIP)/ThioMac-CEBPb-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 1 |
| Score: | 0.60
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | GGTTGGACCA--
--TTGCAACATN |
|


|
|
POL011.1_XCPE1/Jaspar
| Match Rank: | 2 |
| Score: | 0.60
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GGTTGGACCA
GGGCGGGACC- |
|


|
|
GLI3(Zf)/GLI3-ChIP-Chip(GSE11077)/Homer
| Match Rank: | 3 |
| Score: | 0.58
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---GGTTGGACCA
CGTGGGTGGTCC- |
|


|
|
Atf4(bZIP)/MEF-Atf4-ChIP-Seq(GSE35681)/Homer
| Match Rank: | 4 |
| Score: | 0.58
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | GGTTGGACCA-
-ATTGCATCAK |
|


|
|
Chop(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer
| Match Rank: | 5 |
| Score: | 0.57
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | GGTTGGACCA-
-ATTGCATCAT |
|


|
|
MA0133.1_BRCA1/Jaspar
| Match Rank: | 6 |
| Score: | 0.57
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -GGTTGGACCA
GTGTTGN---- |
|

|
|
PB0196.1_Zbtb7b_2/Jaspar
| Match Rank: | 7 |
| Score: | 0.56
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GGTTGGACCA-------
CATAAGACCACCATTAC |
|


|
|
MA0466.1_CEBPB/Jaspar
| Match Rank: | 8 |
| Score: | 0.56
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GGTTGGACCA-
TATTGCACAAT |
|


|
|
PB0029.1_Hic1_1/Jaspar
| Match Rank: | 9 |
| Score: | 0.54
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----GGTTGGACCA--
NGTAGGTTGGCATNNN |
|


|
|
MA0152.1_NFATC2/Jaspar
| Match Rank: | 10 |
| Score: | 0.54
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | GGTTGGACCA
---TGGAAAA |
|


|
|





















