| p-value: | 1e-11 |
| log p-value: | -2.705e+01 |
| Information Content per bp: | 1.590 |
| Number of Target Sequences with motif | 17.0 |
| Percentage of Target Sequences with motif | 1.70% |
| Number of Background Sequences with motif | 4.4 |
| Percentage of Background Sequences with motif | 0.18% |
| Average Position of motif in Targets | 332.4 +/- 352.7bp |
| Average Position of motif in Background | 306.0 +/- 295.8bp |
| Strand Bias (log2 ratio + to - strand density) | 1.3 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PB0174.1_Sox30_2/Jaspar
| Match Rank: | 1 |
| Score: | 0.62
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | TATGATGGTAAT----
TAAGATTATAATACGG |
|


|
|
PB0064.1_Sox14_1/Jaspar
| Match Rank: | 2 |
| Score: | 0.60
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TATGATGGTAAT----
NNTAATTATAATTNNN |
|


|
|
PB0069.1_Sox21_1/Jaspar
| Match Rank: | 3 |
| Score: | 0.60
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | TATGATGGTAAT----
TTTAATTATAATTAAG |
|


|
|
GATA-IR4(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer
| Match Rank: | 4 |
| Score: | 0.60
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | TATGATGGTAAT----
-NAGATWNBNATCTNN |
|


|
|
OCT4-SOX2-TCF-NANOG((POU/Homeobox/HMG)/mES-Oct4-ChIP-Seq(GSE11431)/Homer
| Match Rank: | 5 |
| Score: | 0.60
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---TATGATGGTAAT
CATTGTTATGCAAAT |
|


|
|
PB0079.1_Sry_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.59
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TATGATGGTAAT----
NANTATTATAATTNNN |
|


|
|
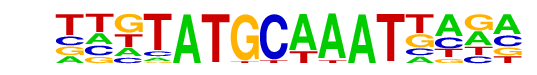
PH0145.1_Pou2f3/Jaspar
| Match Rank: | 7 |
| Score: | 0.58
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | TATGATGGTAAT----
TTGTATGCAAATTAGA |
|

|
|
PB0172.1_Sox1_2/Jaspar
| Match Rank: | 8 |
| Score: | 0.58
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -TATGATGGTAAT--
CTATAATTGTTAGCG |
|


|
|
Oct4(POU/Homeobox)/mES-Oct4-ChIP-Seq(GSE11431)/Homer
| Match Rank: | 9 |
| Score: | 0.57
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | TATGATGGTAAT
--TTATGCAAAT |
|


|
|
MA0142.1_Pou5f1::Sox2/Jaspar
| Match Rank: | 10 |
| Score: | 0.57
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---TATGATGGTAAT
CTTTGTTATGCAAAT |
|


|
|





















