| p-value: | 1e-16 |
| log p-value: | -3.783e+01 |
| Information Content per bp: | 1.963 |
| Number of Target Sequences with motif | 47.0 |
| Percentage of Target Sequences with motif | 1.88% |
| Number of Background Sequences with motif | 4.3 |
| Percentage of Background Sequences with motif | 0.44% |
| Average Position of motif in Targets | 439.0 +/- 339.6bp |
| Average Position of motif in Background | 521.4 +/- 355.5bp |
| Strand Bias (log2 ratio + to - strand density) | -0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.04 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
MA0089.1_NFE2L1::MafG/Jaspar
| Match Rank: | 1 |
| Score: | 0.79
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | ACATGACC
-CATGAC- |
|


|
|
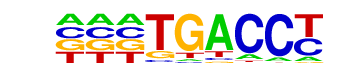
MF0004.1_Nuclear_Receptor_class/Jaspar
| Match Rank: | 2 |
| Score: | 0.77
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | ACATGACC-
---TGACCT |
|

|
|
MA0093.2_USF1/Jaspar
| Match Rank: | 3 |
| Score: | 0.76
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---ACATGACC
GCCACGTGACC |
|


|
|
E-box(HLH)/Promoter/Homer
| Match Rank: | 4 |
| Score: | 0.74
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --ACATGACC--
TCACGTGACCGG |
|


|
|
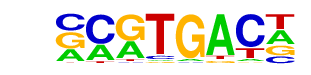
MA0067.1_Pax2/Jaspar
| Match Rank: | 5 |
| Score: | 0.74
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | ACATGACC
NCGTGACN |
|

|
|
PB0157.1_Rara_2/Jaspar
| Match Rank: | 6 |
| Score: | 0.72
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -ACATGACC-------
NNCNTGACCCCGCTCT |
|


|
|
PB0057.1_Rxra_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.72
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --ACATGACC-------
TGTCGTGACCCCTTAAT |
|


|
|
MA0071.1_RORA_1/Jaspar
| Match Rank: | 8 |
| Score: | 0.71
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | ACATGACC-----
---TGACCTTGAT |
|


|
|
PB0049.1_Nr2f2_1/Jaspar
| Match Rank: | 9 |
| Score: | 0.71
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -ACATGACC-------
NNNNTGACCTTTNNNN |
|


|
|
PB0153.1_Nr2f2_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.70
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -ACATGACC-------
NNNNTGACCCGGCGCG |
|


|
|





















