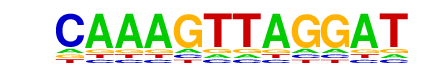
| p-value: | 1e-10 |
| log p-value: | -2.521e+01 |
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 13.0 |
| Percentage of Target Sequences with motif | 1.30% |
| Number of Background Sequences with motif | 3.0 |
| Percentage of Background Sequences with motif | 0.13% |
| Average Position of motif in Targets | 361.8 +/- 292.7bp |
| Average Position of motif in Background | 434.6 +/- 140.8bp |
| Strand Bias (log2 ratio + to - strand density) | 0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
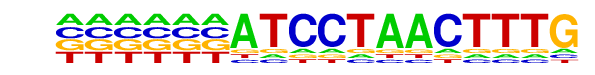
PB0181.1_Spdef_2/Jaspar
| Match Rank: | 1 |
| Score: | 0.54
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------ATCCTAACTTTG
GATAACATCCTAGTAG-- |
|

|
|
MA0029.1_Mecom/Jaspar
| Match Rank: | 2 |
| Score: | 0.54
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----ATCCTAACTTTG
TNTTATCTTATCTT-- |
|


|
|
MA0442.1_SOX10/Jaspar
| Match Rank: | 3 |
| Score: | 0.53
| | Offset: | 7
| | Orientation: | forward strand |
| Alignment: | ATCCTAACTTTG-
-------CTTTGT |
|


|
|
PB0161.1_Rxra_2/Jaspar
| Match Rank: | 4 |
| Score: | 0.52
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -ATCCTAACTTTG---
NNNNCAACCTTCGNGA |
|


|
|
MA0114.2_HNF4A/Jaspar
| Match Rank: | 5 |
| Score: | 0.52
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | ATCCTAACTTTG-----
--CTGGACTTTGGACTC |
|


|
|
MA0141.2_Esrrb/Jaspar
| Match Rank: | 6 |
| Score: | 0.52
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | ATCCTAACTTTG----
----TGACCTTGANNN |
|


|
|
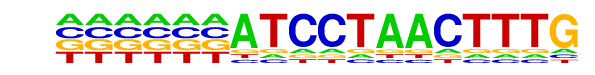
PB0115.1_Ehf_2/Jaspar
| Match Rank: | 7 |
| Score: | 0.52
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------ATCCTAACTTTG
TAGTATTTCCGATCTT-- |
|

|
|
Nr5a2(NR)/Pancreas-LRH1-ChIP-Seq(GSE34295)/Homer
| Match Rank: | 8 |
| Score: | 0.51
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | ATCCTAACTTTG--
----TGACCTTGAV |
|


|
|
MA0484.1_HNF4G/Jaspar
| Match Rank: | 9 |
| Score: | 0.51
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | ATCCTAACTTTG------
---TGGACTTTGNNCTCN |
|


|
|
PB0121.1_Foxj3_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.51
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---ATCCTAACTTTG--
AACACCAAAACAAAGGA |
|


|
|