| p-value: | 1e-10 |
| log p-value: | -2.365e+01 |
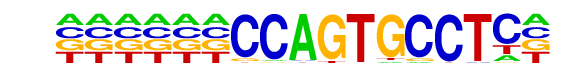
| Information Content per bp: | 1.822 |
| Number of Target Sequences with motif | 52.0 |
| Percentage of Target Sequences with motif | 5.21% |
| Number of Background Sequences with motif | 42.9 |
| Percentage of Background Sequences with motif | 1.87% |
| Average Position of motif in Targets | 392.1 +/- 282.0bp |
| Average Position of motif in Background | 468.9 +/- 344.9bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.04 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PB0195.1_Zbtb3_2/Jaspar
| Match Rank: | 1 |
| Score: | 0.66
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------CCAGTGCCTC
NNNNTGCCAGTGATTG |
|


|
|
PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 2 |
| Score: | 0.64
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CCAGTGCCTC-
-CACTTCCTCT |
|


|
|
MA0080.3_Spi1/Jaspar
| Match Rank: | 3 |
| Score: | 0.61
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CCAGTGCCTC-----
NCACTTCCTCTTTTN |
|


|
|
PB0091.1_Zbtb3_1/Jaspar
| Match Rank: | 4 |
| Score: | 0.61
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------CCAGTGCCTC-
NNNANTGCAGTGCNNTT |
|

|
|
ETS:RUNX/Jurkat-RUNX1-ChIP-Seq(GSE17954)/Homer
| Match Rank: | 5 |
| Score: | 0.61
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CCAGTGCCTC-
ACCACATCCTGT |
|


|
|
PH0171.1_Nkx2-1/Jaspar
| Match Rank: | 6 |
| Score: | 0.58
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----CCAGTGCCTC-
AANTTCAAGTGGCTTN |
|


|
|
Nkx3.1(Homeobox)/LNCaP-Nkx3.1-ChIP-Seq(GSE28264)/Homer
| Match Rank: | 7 |
| Score: | 0.58
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CCAGTGCCTC
TTAAGTGCTT- |
|


|
|
MA0146.2_Zfx/Jaspar
| Match Rank: | 8 |
| Score: | 0.57
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----CCAGTGCCTC
GGGGCCGAGGCCTG |
|


|
|
MA0474.1_Erg/Jaspar
| Match Rank: | 9 |
| Score: | 0.57
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CCAGTGCCTC-
CCACTTCCTGT |
|


|
|
Nkx2.5(Homeobox)/HL1-Nkx2.5.biotin-ChIP-Seq(GSE21529)/Homer
| Match Rank: | 10 |
| Score: | 0.56
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CCAGTGCCTC
TTGAGTGSTT- |
|


|
|





















