| p-value: | 1e-12 |
| log p-value: | -2.770e+01 |
| Information Content per bp: | 1.825 |
| Number of Target Sequences with motif | 54.0 |
| Percentage of Target Sequences with motif | 5.41% |
| Number of Background Sequences with motif | 40.5 |
| Percentage of Background Sequences with motif | 1.77% |
| Average Position of motif in Targets | 382.6 +/- 311.9bp |
| Average Position of motif in Background | 508.7 +/- 410.5bp |
| Strand Bias (log2 ratio + to - strand density) | -0.4 |
| Multiplicity (# of sites on avg that occur together) | 1.04 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
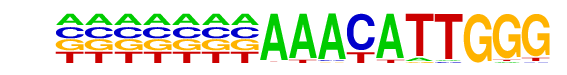
Sox2(HMG)/mES-Sox2-ChIP-Seq(GSE11431)/Homer
| Match Rank: | 1 |
| Score: | 0.68
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | AAACATTGGG
GAACAATGGN |
|


|
|
MA0077.1_SOX9/Jaspar
| Match Rank: | 2 |
| Score: | 0.68
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | AAACATTGGG
GAACAATGG- |
|


|
|
MA0515.1_Sox6/Jaspar
| Match Rank: | 3 |
| Score: | 0.66
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -AAACATTGGG
AAAACAATGG- |
|


|
|
PB0132.1_Hbp1_2/Jaspar
| Match Rank: | 4 |
| Score: | 0.66
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---AAACATTGGG----
NNTNNACAATGGGANNN |
|


|
|
MA0143.3_Sox2/Jaspar
| Match Rank: | 5 |
| Score: | 0.66
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | AAACATTGGG
-AACAAAGG- |
|


|
|
MA0040.1_Foxq1/Jaspar
| Match Rank: | 6 |
| Score: | 0.64
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---AAACATTGGG
AATAAACAATN-- |
|


|
|
PB0002.1_Arid5a_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.64
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----AAACATTGGG
NNTNNCAATATTAG- |
|


|
|
PB0121.1_Foxj3_2/Jaspar
| Match Rank: | 8 |
| Score: | 0.62
| | Offset: | -7
| | Orientation: | forward strand |
| Alignment: | -------AAACATTGGG
AACACCAAAACAAAGGA |
|

|
|
MF0005.1_Forkhead_class/Jaspar
| Match Rank: | 9 |
| Score: | 0.61
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----AAACATTGGG
AAATAAACA----- |
|


|
|
MA0153.1_HNF1B/Jaspar
| Match Rank: | 10 |
| Score: | 0.61
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---AAACATTGGG
GTTAAATATTAA- |
|


|
|





















