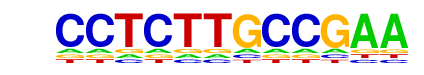
| p-value: | 1e-15 |
| log p-value: | -3.558e+01 |
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 24.0 |
| Percentage of Target Sequences with motif | 0.17% |
| Number of Background Sequences with motif | 3.0 |
| Percentage of Background Sequences with motif | 0.03% |
| Average Position of motif in Targets | 541.6 +/- 441.3bp |
| Average Position of motif in Background | 685.7 +/- 442.9bp |
| Strand Bias (log2 ratio + to - strand density) | 1.2 |
| Multiplicity (# of sites on avg that occur together) | 1.08 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
SD0002.1_at_AC_acceptor/Jaspar
| Match Rank: | 1 |
| Score: | 0.55
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CCTCTTGCCGAA
NNACTTGCCTT- |
|
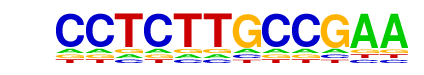

|
|
PPARE(NR/DR1)/3T3L1-Pparg-ChIP-Seq(GSE13511)/Homer
| Match Rank: | 2 |
| Score: | 0.53
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---CCTCTTGCCGAA
TGACCTTTGCCCCA- |
|


|
|
MA0102.3_CEBPA/Jaspar
| Match Rank: | 3 |
| Score: | 0.53
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | CCTCTTGCCGAA--
---ATTGCACAATA |
|
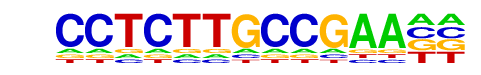

|
|
MA0065.2_PPARG::RXRA/Jaspar
| Match Rank: | 4 |
| Score: | 0.52
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---CCTCTTGCCGAA
TGACCTTTGCCCTAN |
|


|
|
RXR(NR/DR1)/3T3L1-RXR-ChIP-Seq(GSE13511)/Homer
| Match Rank: | 5 |
| Score: | 0.52
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---CCTCTTGCCGAA
TGACCTTTGCCCTA- |
|


|
|
MA0466.1_CEBPB/Jaspar
| Match Rank: | 6 |
| Score: | 0.52
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CCTCTTGCCGAA-
--TATTGCACAAT |
|
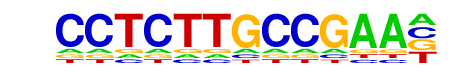

|
|
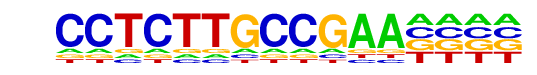
PB0145.1_Mafb_2/Jaspar
| Match Rank: | 7 |
| Score: | 0.51
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CCTCTTGCCGAA----
-CAATTGCAAAAATAT |
|

|
|
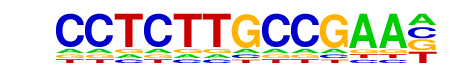
CEBP(bZIP)/CEBPb-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 8 |
| Score: | 0.50
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | CCTCTTGCCGAA-
---GTTGCGCAAT |
|

|
|
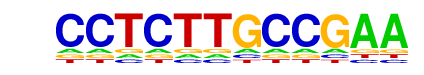
NF1-halfsite(CTF)/LNCaP-NF1-ChIP-Seq(Unpublished)/Homer
| Match Rank: | 9 |
| Score: | 0.50
| | Offset: | 4
| | Orientation: | forward strand |
| Alignment: | CCTCTTGCCGAA
----TTGCCAAG |
|

|
|
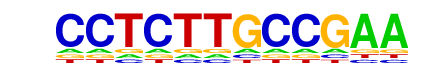
Unknown-ESC-element/mES-Nanog-ChIP-Seq(GSE11724)/Homer
| Match Rank: | 10 |
| Score: | 0.49
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CCTCTTGCCGAA
CCCCCTGCTGTG |
|

|
|