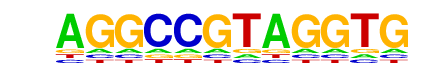
| p-value: | 1e-15 |
| log p-value: | -3.558e+01 |
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 24.0 |
| Percentage of Target Sequences with motif | 0.17% |
| Number of Background Sequences with motif | 2.1 |
| Percentage of Background Sequences with motif | 0.02% |
| Average Position of motif in Targets | 448.4 +/- 413.4bp |
| Average Position of motif in Background | 243.5 +/- 27.5bp |
| Strand Bias (log2 ratio + to - strand density) | 1.3 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
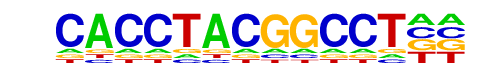
ZNF711(Zf)/SH-SY5Y-ZNF711-ChIP-Seq(GSE20673)/Homer
| Match Rank: | 1 |
| Score: | 0.59
| | Offset: | 6
| | Orientation: | forward strand |
| Alignment: | CACCTACGGCCT--
------AGGCCTAG |
|

|
|
MA0146.2_Zfx/Jaspar
| Match Rank: | 2 |
| Score: | 0.58
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CACCTACGGCCT-
GGGGCCGAGGCCTG |
|


|
|
Tbx5(T-box)/HL1-Tbx5.biotin-ChIP-Seq(GSE21529)/Homer
| Match Rank: | 3 |
| Score: | 0.58
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---CACCTACGGCCT
TGACACCT------- |
|


|
|
PB0150.1_Mybl1_2/Jaspar
| Match Rank: | 4 |
| Score: | 0.57
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CACCTACGGCCT--
CGACCAACTGCCGTG |
|


|
|
MA0009.1_T/Jaspar
| Match Rank: | 5 |
| Score: | 0.56
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----CACCTACGGCCT
TTCACACCTAG----- |
|


|
|
PB0189.1_Tcfap2a_2/Jaspar
| Match Rank: | 6 |
| Score: | 0.56
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CACCTACGGCCT-
TCACCTCTGGGCAG |
|


|
|
E2A-nearPU.1(HLH)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 7 |
| Score: | 0.56
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CACCTACGGCCT
NNCACCTGNN---- |
|


|
|
PB0117.1_Eomes_2/Jaspar
| Match Rank: | 8 |
| Score: | 0.56
| | Offset: | -7
| | Orientation: | reverse strand |
| Alignment: | -------CACCTACGGCCT
NNGGCGACACCTCNNN--- |
|


|
|
MA0163.1_PLAG1/Jaspar
| Match Rank: | 9 |
| Score: | 0.55
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CACCTACGGCCT-
CCCCCTTGGGCCCC |
|


|
|
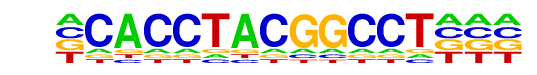
PB0149.1_Myb_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.55
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CACCTACGGCCT---
CGACCAACTGCCATGC |
|

|
|